new_mean <- function(x) {
sum(x) / length(x)
}Descriptive Stats & Distributions
http://tinyurl.com/4dfuycvt
Descriptive stats

Mean & Median
Sample mean: \[ \overline{x} = \frac{1}{n}\sum^{n}_{i=1}x_{i} \]
You can create the function:
Sample median: \[ median= \begin{cases} x_{(n+1)/2} &\text{if $n \equiv 1$} \\ \frac{(x_{n/2} + x_{((n/2)+1)})}{2} &\text{if $n \equiv 0$} \end{cases} \]
In R a freshly defined function:
new_median <- function(x) {
if (length(x) %% 2 != 0) {
sort(x)[ceiling(length(x) / 2)]
} else {
(sort(x)[length(x) / 2] + sort(x)[(length(x) / 2) + 1]) / 2
}
}- 1
-
The expression
length(x) %% 2 != 0checks whether the sample size is odd (impar). The modulus operator%%returns the remainder oflength(x)divided by 2; if the remainder is 1 (non-zero), the condition is TRUE and the function uses the odd-case formula. - 2
- Equation for n if it is odd (impar)
- 3
- Equation for n if it is even (par)
Variance, Standard deviation & Variation coefficient
Sample variance (unbiased): \[ S^{2} = \frac{1}{n-1}\sum^{n}_{i=1}(x_{i}-\overline{x})^{2} \]
Standard deviation
\[ S = \sqrt{\frac{1}{n-1}\sum^{n}_{i=1}(x_{i}-\overline{x})^{2}} \]
Variation coefficient
\[ CV = \frac{S}{\overline{x}} \times 100 \]
Confidence intervals
Estimates the range within which a population parameter is likely to fall. It provides a way to express the uncertainty or margin of error associated with a sample estimate. \[ \text{IC} = \bar{x} \pm Z \left( \frac{\sigma}{\sqrt{n}} \right) \]
Or if the population standard deviation of the population is unknown:
\[ \text{IC} = \bar{x} \pm t \left( \frac{s}{\sqrt{n}} \right) \]
Using dplyr for summarizing stats
How to compute all summary statistics for all numeric variables in a dataset?
Solution in the next slide, but before take your time, breath and go back to previous slide…
Distributions
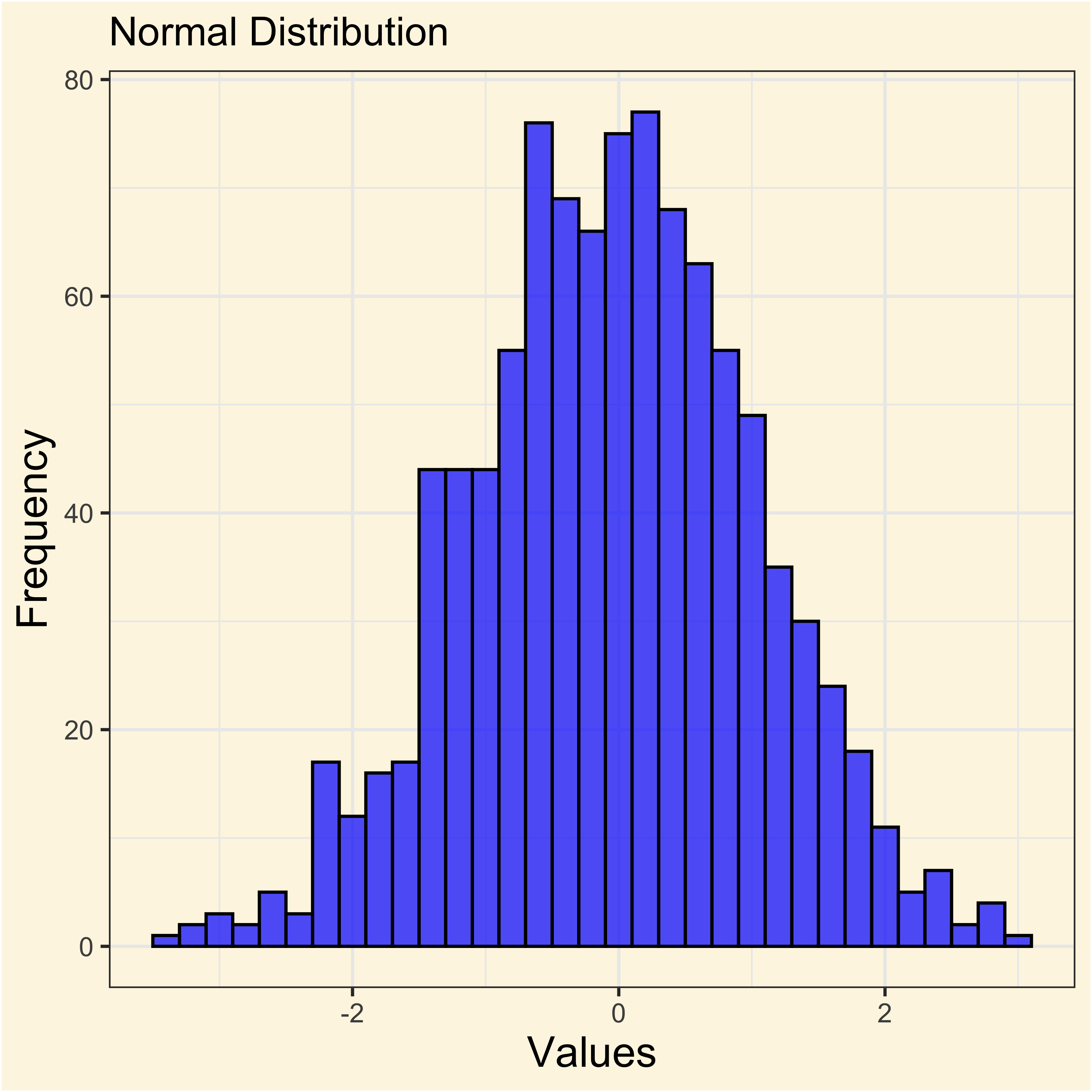
The standard normal distribution
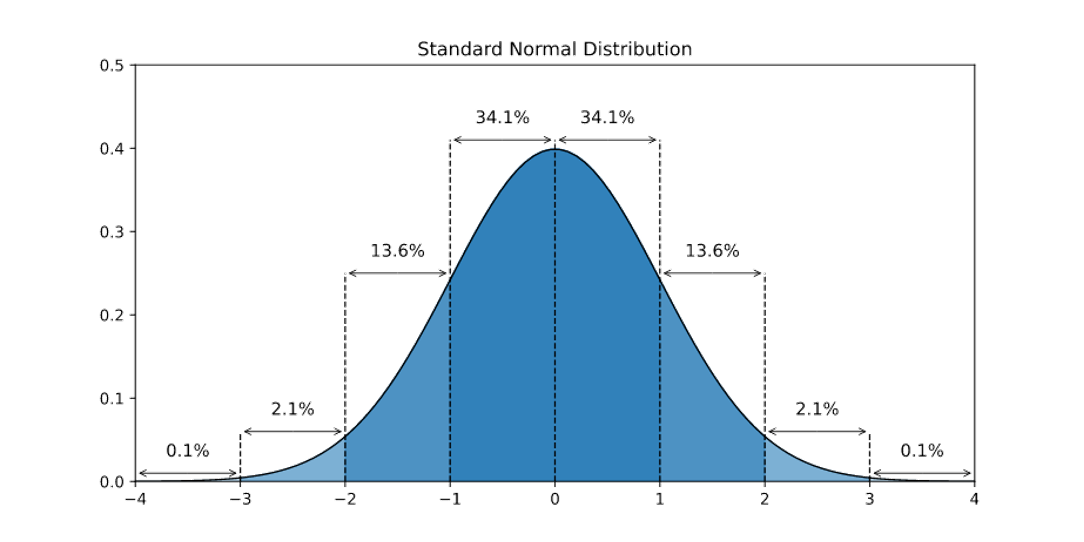
Where \(z = \frac{x - \mu} {\sigma}\) is the standardization function, the result is that \(\sigma = 1\) and \(\mu = 0\)
family of normality functions in R
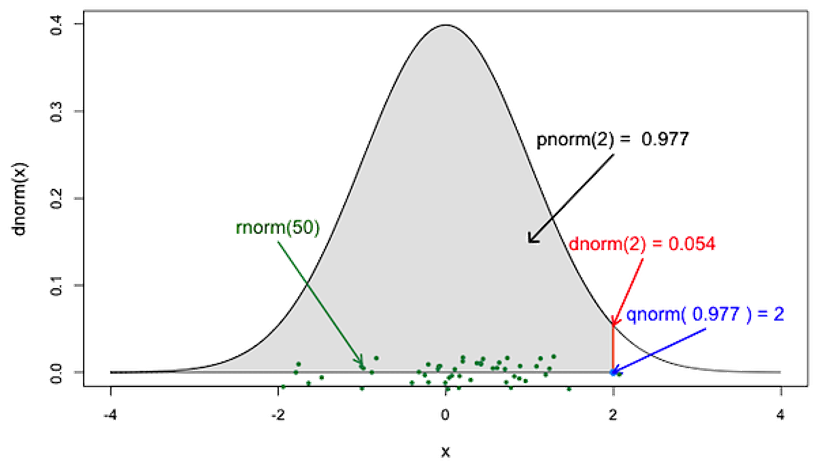
rnorm()generates pseudorandom normal numbers.dnorm()is the probability density function (PDF)pnorm()is the cumulative density functionqnorm()calculates the quantile of the normal distribution
Let’s try with an example:
Now, the Adelie penguins display an average bill length of 38.8 mm and its standard variation is 2.6 mm. What’s the percentage of penguins of 40 mm or smaller?
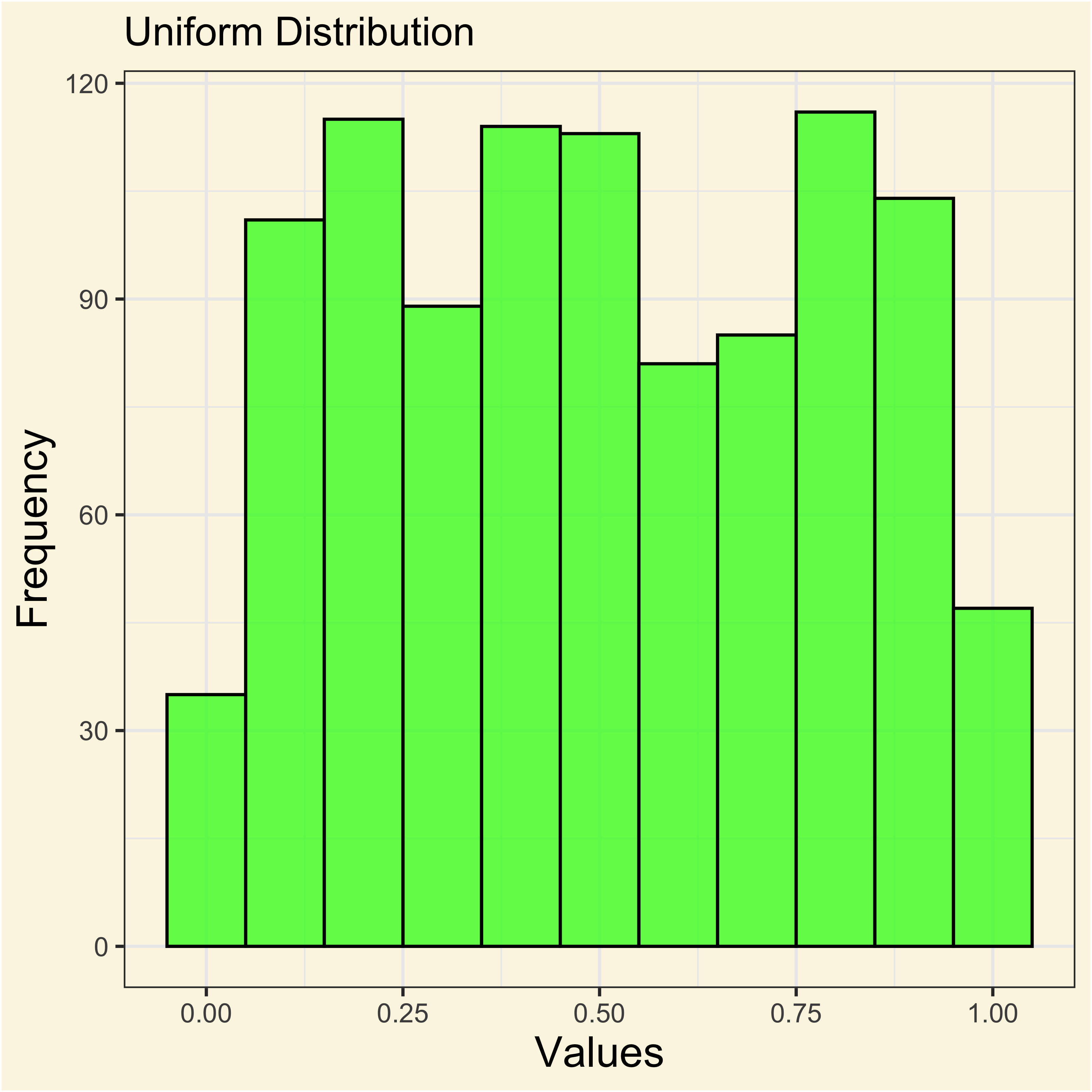
[1] 0.6777938When the frequencies of a random variable \(X\) cluster around a central value, it is said that it follows a normal distribution.
In summary a variable that appears to follow a normal distribution displays three properties:
- Most values clustered around the average.
- Extreme values are less frequent, but not impossibles.
- Distribution display are quite symmetric from the mean.
BIOL2205 - Inferencia e Informática - DCB - Uniandes