t.test(body_mass_g ~ sex, adelie_penguins)Parametric & Non-Parametric tests
https://bit.ly/3V7CMsZ
How do we test whether two samples are different?
Do male penguins display greater body sizes than female?
adelie_penguins <- penguins |>
filter(species == "Adelie") |>
drop_na()
ggplot(adelie_penguins, aes(x = sex, y = body_mass_g, fill = sex)) +
geom_boxplot(alpha = 0.5) +
geom_jitter(width = 0.2) +
labs(
y = "Body mass (g)",
x = ""
)
Let’s try to test the resulting hypotheses:
Null hypothesis: \[H_{0}: \mu_{1} = \mu_{2}\] No variation between means
Alternative hypothesis: \[H_{A}: \mu_{1} \neq \mu_{2}\] Variation between means


Let’s make some important assumptions
- The two samples are independent.
- Variables display normal distribution.
- The two samples display equal variances.
Then,

And,

The T Distribution
\[ \bar{X}_{1} - \bar{X}_{2} \sim N\Bigg[\mu_{1} - \mu_{2},s^{2}\bigg(\frac{1}{n_{1}}+\frac{1}{n_{2}}\bigg)\Bigg] \]
If \(H_{0}\) is true \(\rightarrow\) \(\mu_{1} - \mu_{2} = 0\), then
\[ \bar{X}_{1} - \bar{X}_{2} \sim N\Bigg[0,s^{2}\bigg(\frac{1}{n_{1}}+\frac{1}{n_{2}}\bigg)\Bigg] \]
\[ \frac{\bar{X}_{1} - \bar{X}_{2}}{s\sqrt{\frac{1}{n_{1}}+\frac{1}{n_{2}}}} \sim N(0,1) \]
\[ \frac{\bar{X}_{1} - \bar{X}_{2}}{s\sqrt{\frac{1}{n_{1}}+\frac{1}{n_{2}}}} = t \]
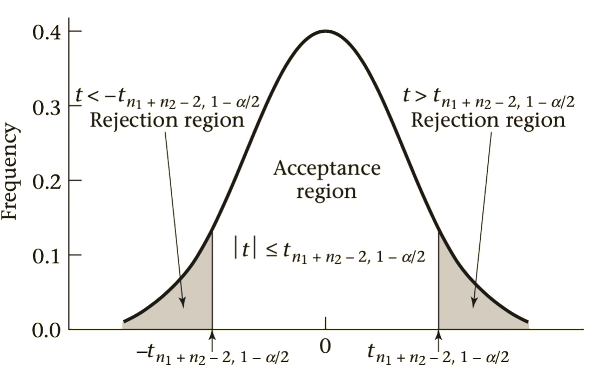
Testing the hypothesis in R
The parametric T-test
A way to test the hypothesis is using the t.test() function:
Welch Two Sample t-test
data: body_mass_g by sex
t = -13.126, df = 135.69, p-value < 2.2e-16
alternative hypothesis: true difference in means between group female and group male is not equal to 0
95 percent confidence interval:
-776.3012 -573.0139
sample estimates:
mean in group female mean in group male
3368.836 4043.493 Another alternative is to use the statsExpressions library:

library(statsExpressions)
adelie_ttest_table <- two_sample_test(
data = adelie_penguins,
x = sex,
y = body_mass_g,
type = "p",
paired = FALSE
)
adelie_ttest_tableAdding stats to the barplot!

Adding comparisons to the plot using ggsignif library!
ggplot(
adelie_penguins,
aes(
x = sex,
y = body_mass_g,
fill = sex
)
) +
geom_boxplot(alpha = 0.5) +
geom_jitter(width = 0.2) +
labs(
y = "Body mass (g)",
x = "",
subtitle = parse(
text = adelie_ttest_table$expression
)
) +
geom_signif(
comparisons = list(c("female", "male")),
test = "t.test",
map_signif_level = TRUE
)
The non-parametric Mann-Whitney/Wilcox test
Let’s make some important assumptions
- The two samples are independent.
- Variables could display non-normal distribution.
- The two samples could display different variances (and sample sizes).
A way to test the hypothesis is using the wilkox.test() function:
wilcox.test(body_mass_g ~ sex, adelie_penguins, paired = FALSE)
Wilcoxon rank sum test with continuity correction
data: body_mass_g by sex
W = 310.5, p-value < 2.2e-16
alternative hypothesis: true location shift is not equal to 0Another alternative is to use the statsExpressions library:
library(statsExpressions)
adelie_wilcox_table <- two_sample_test(
data = adelie_penguins,
x = sex,
y = body_mass_g,
type = "np",
paired = FALSE
)
adelie_wilcox_tableAdding stats to the barplot!
Adding comparisons to the plot using ggsignif library!
ggplot(
adelie_penguins,
aes(
x = sex,
y = body_mass_g,
fill = sex
)
) +
geom_boxplot(alpha = 0.5) +
geom_jitter(width = 0.2) +
labs(
y = "Body mass (g)",
x = "",
subtitle = parse(
text = adelie_wilcox_table$expression
)
) +
geom_signif(
comparisons = list(c("female", "male")),
test = "wilcox.test",
map_signif_level = TRUE
)
BIOL2205 - Inferencia e Informática - DCB - Uniandes

