anovamodel <- aov(data = males, body_mass_g ~ species)ANOVA
https://bit.ly/41mzUK9
Camilo G.
Alejandra S.
Ronald D.
Andrew C.
Mauricio S.
When do we apply ANOVA
When comparing a continuous variable across two or more groups. The question it tries to solve is: “is there a group different from the others?”
Establishing the hypothesis
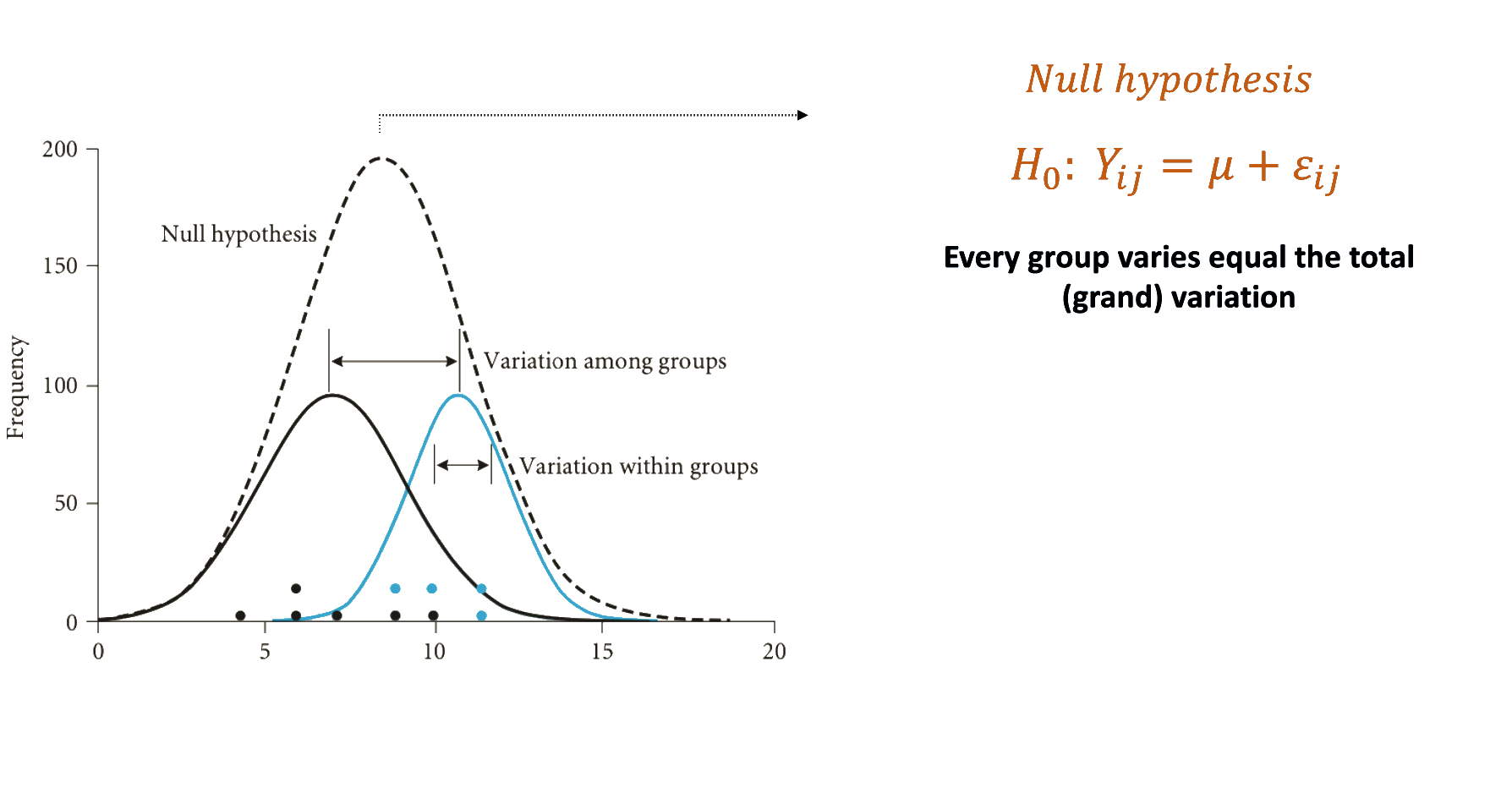
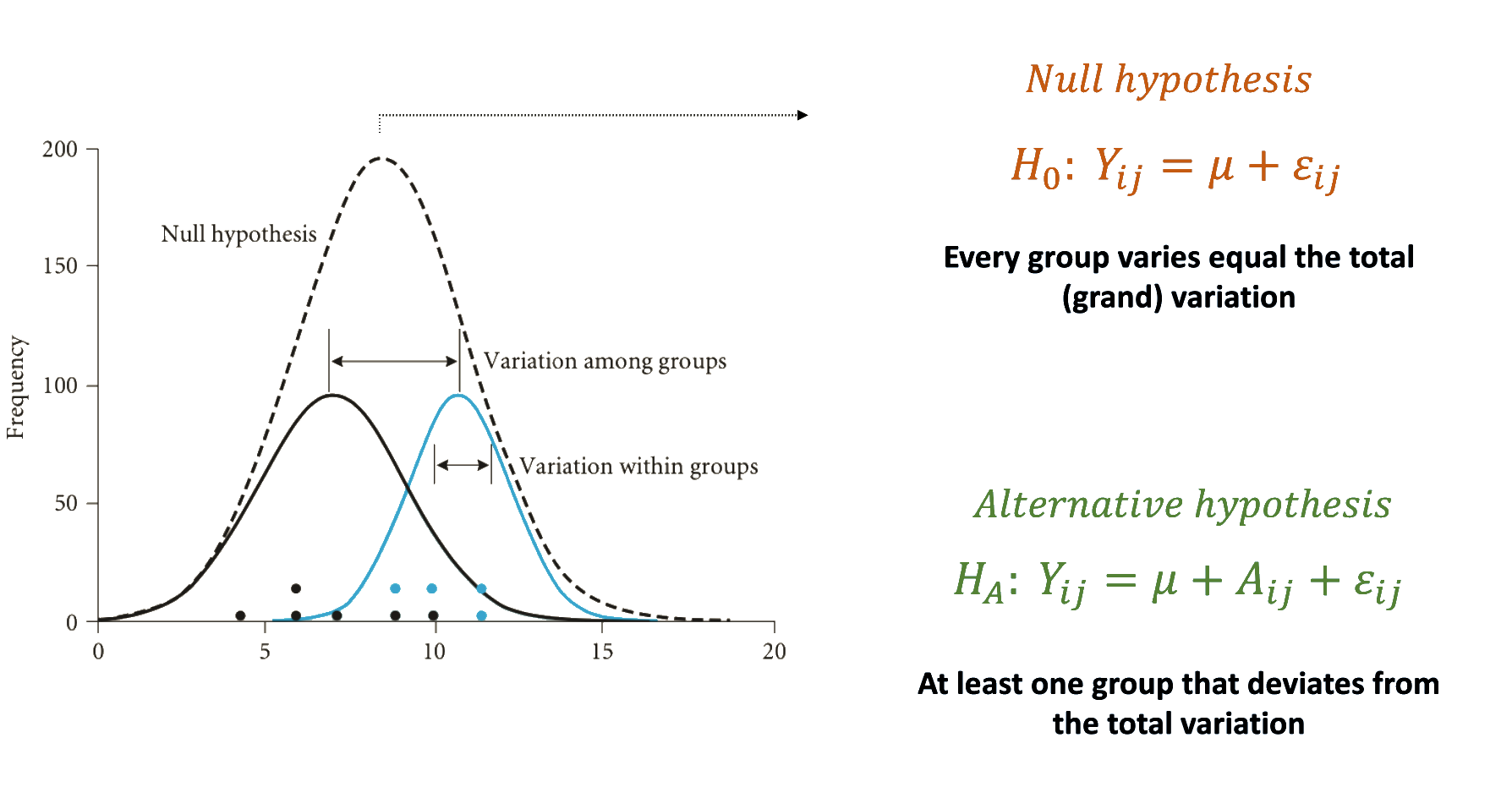
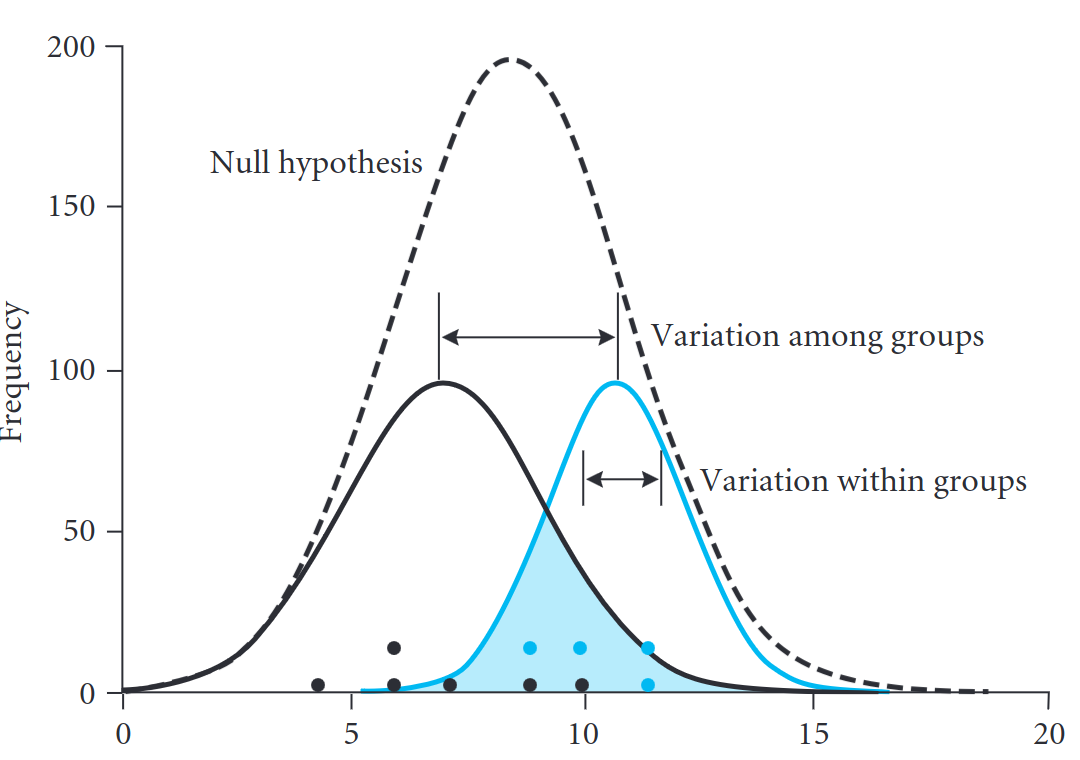
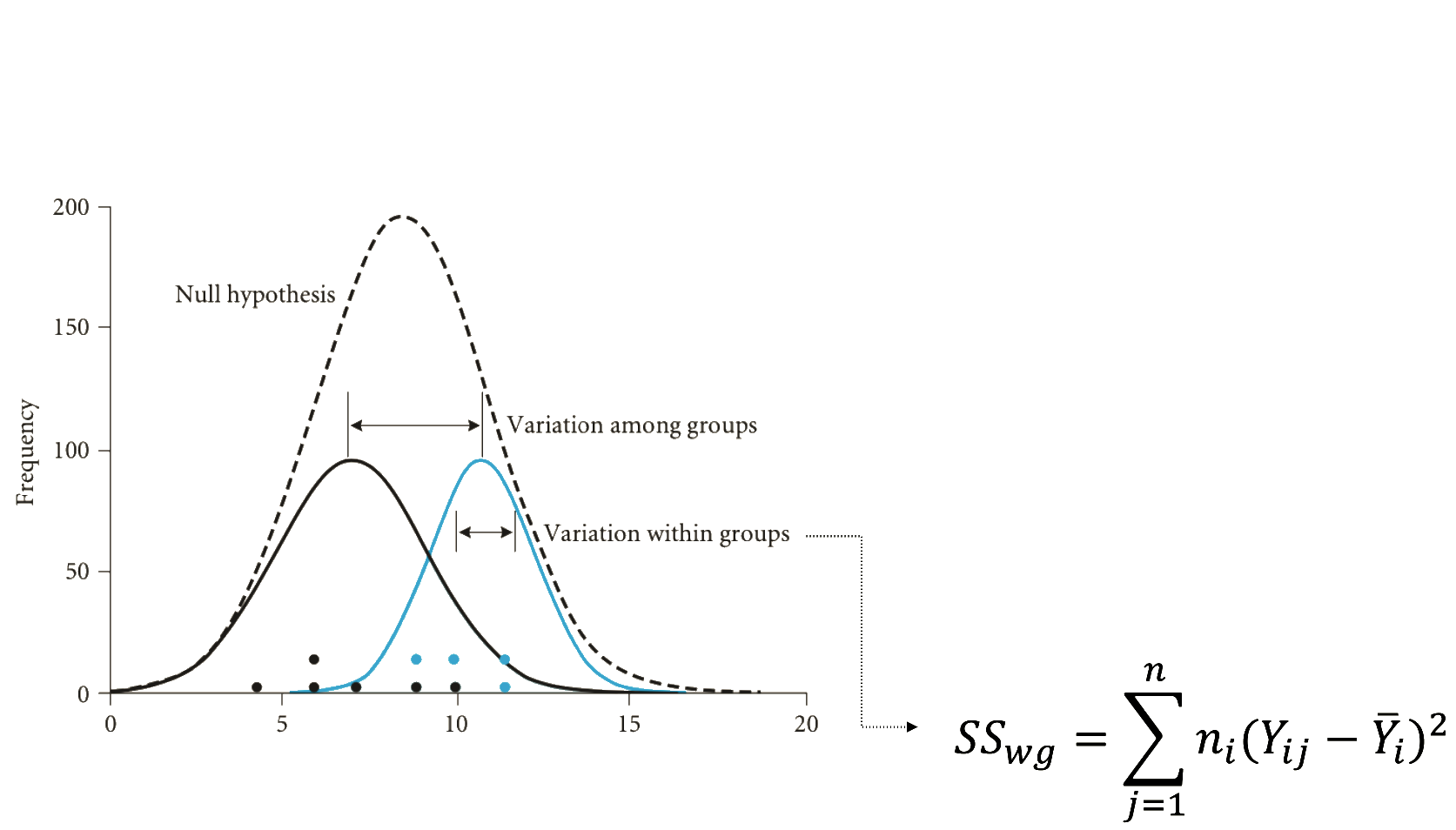
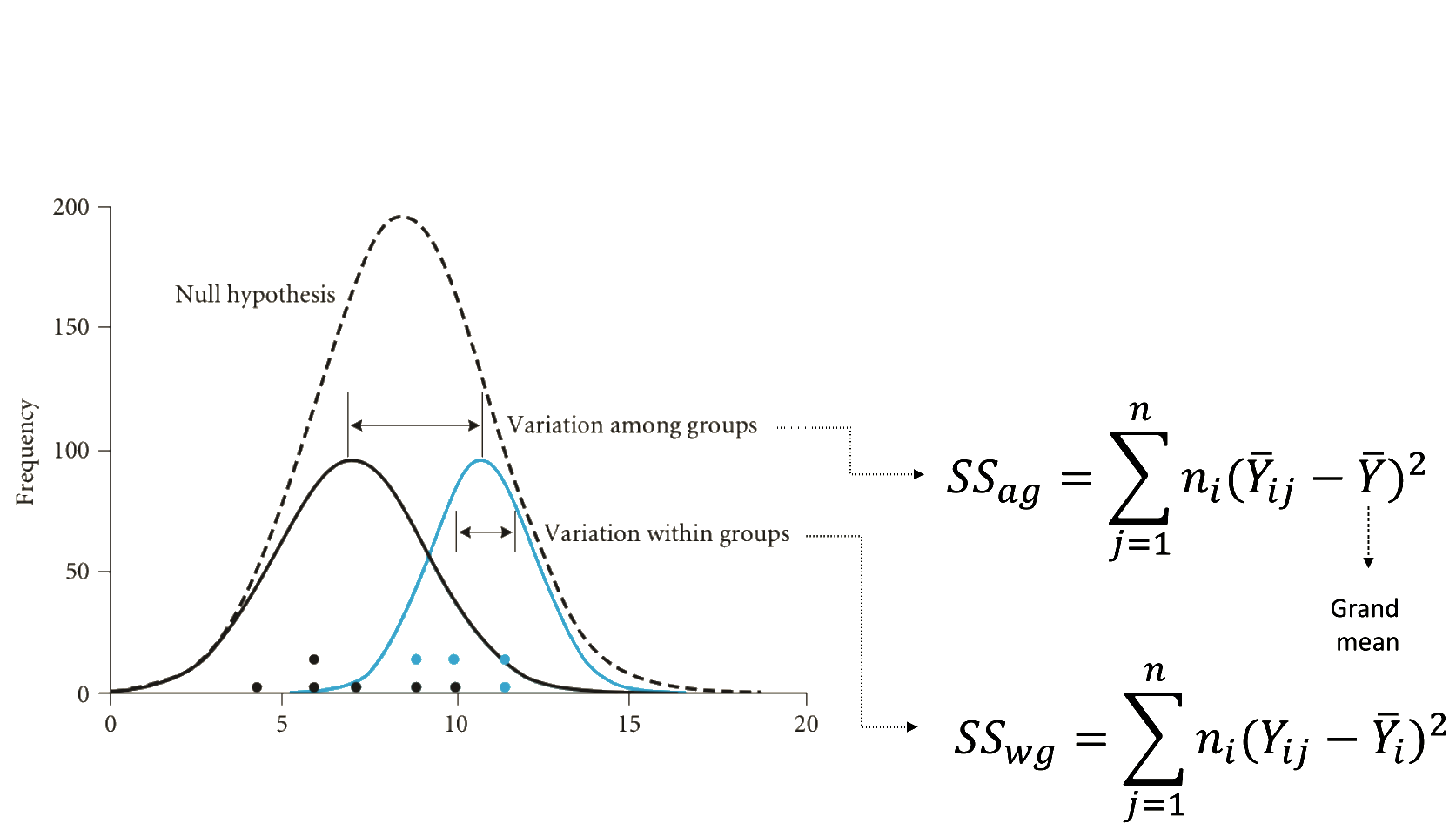
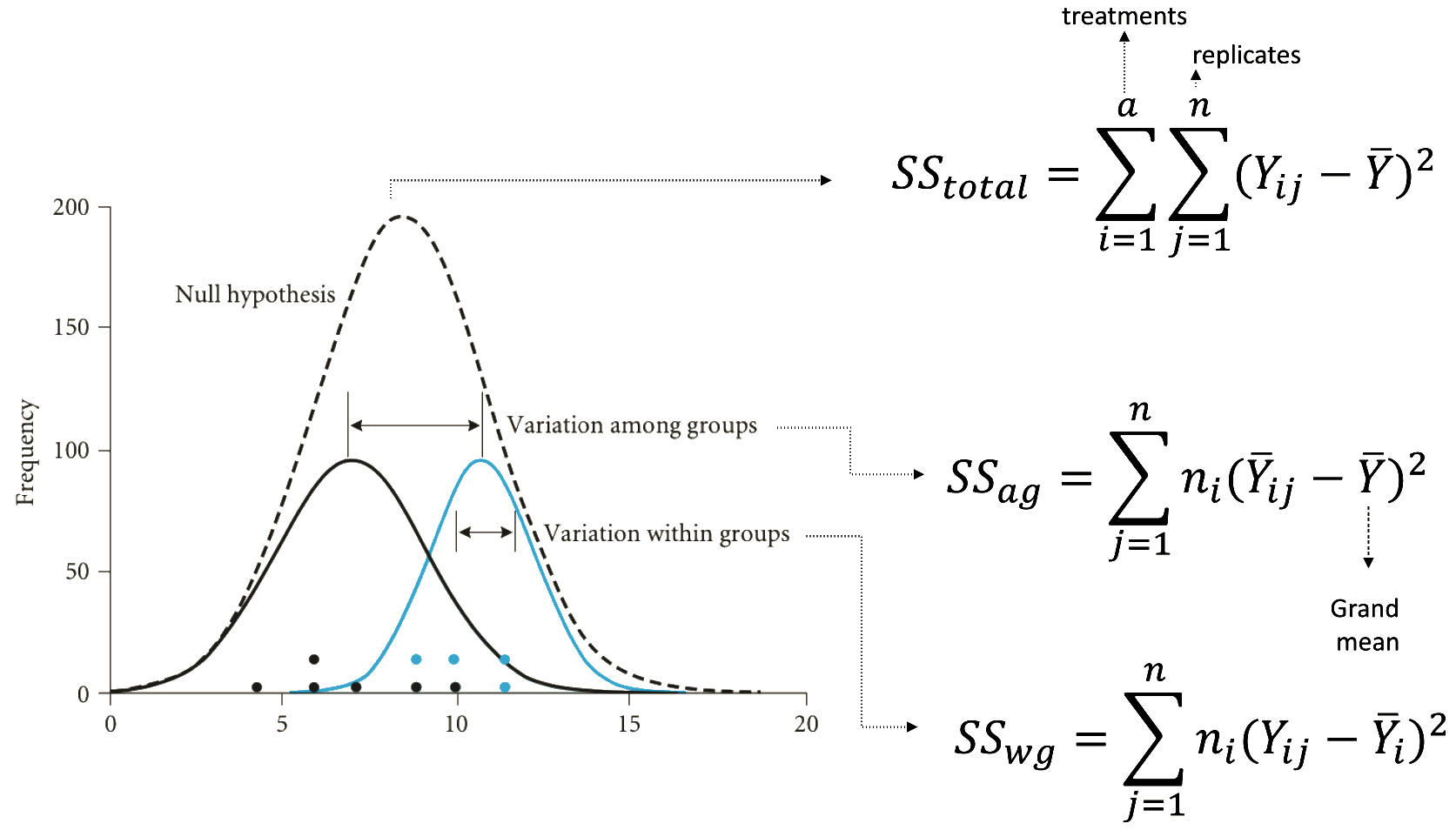
Understanding the fisher distribution
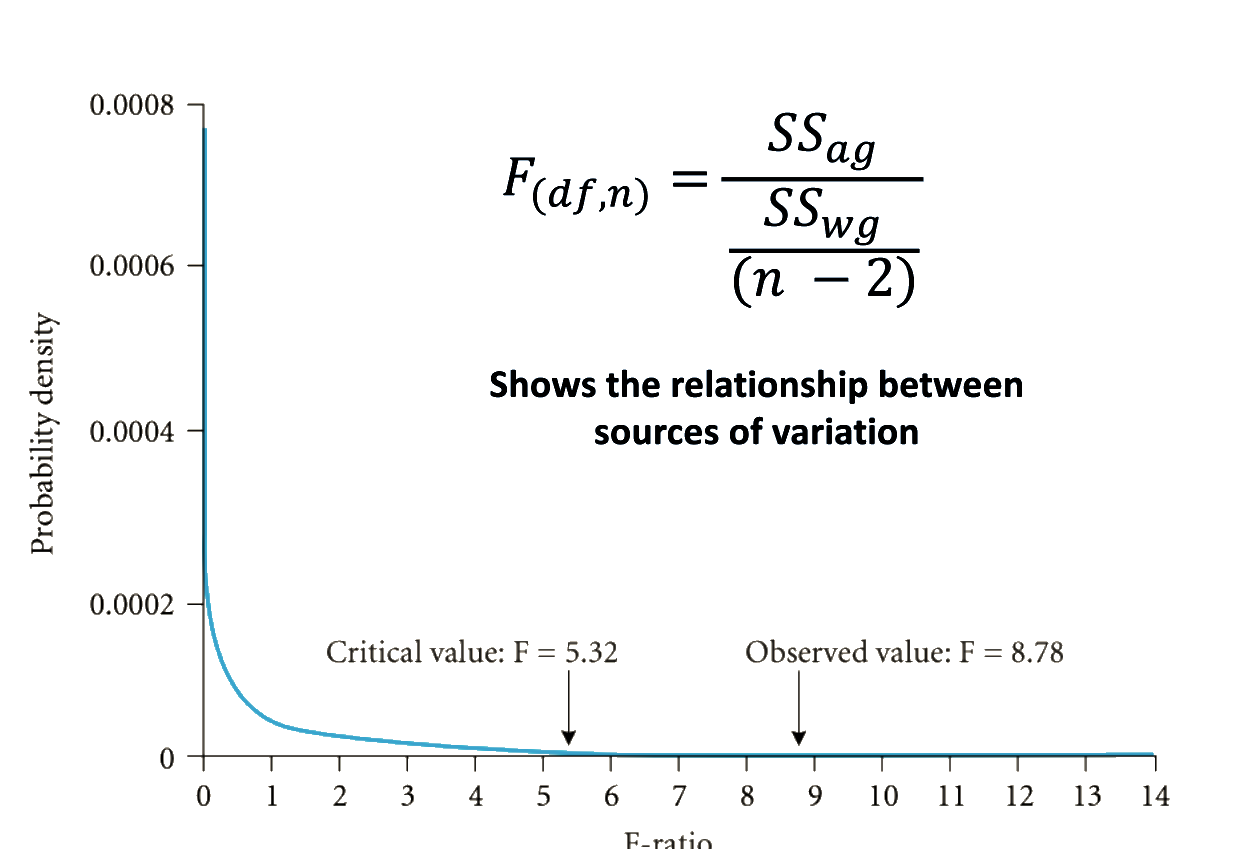

ANOVA assumptions
- Samples are independent (randomly sampled)
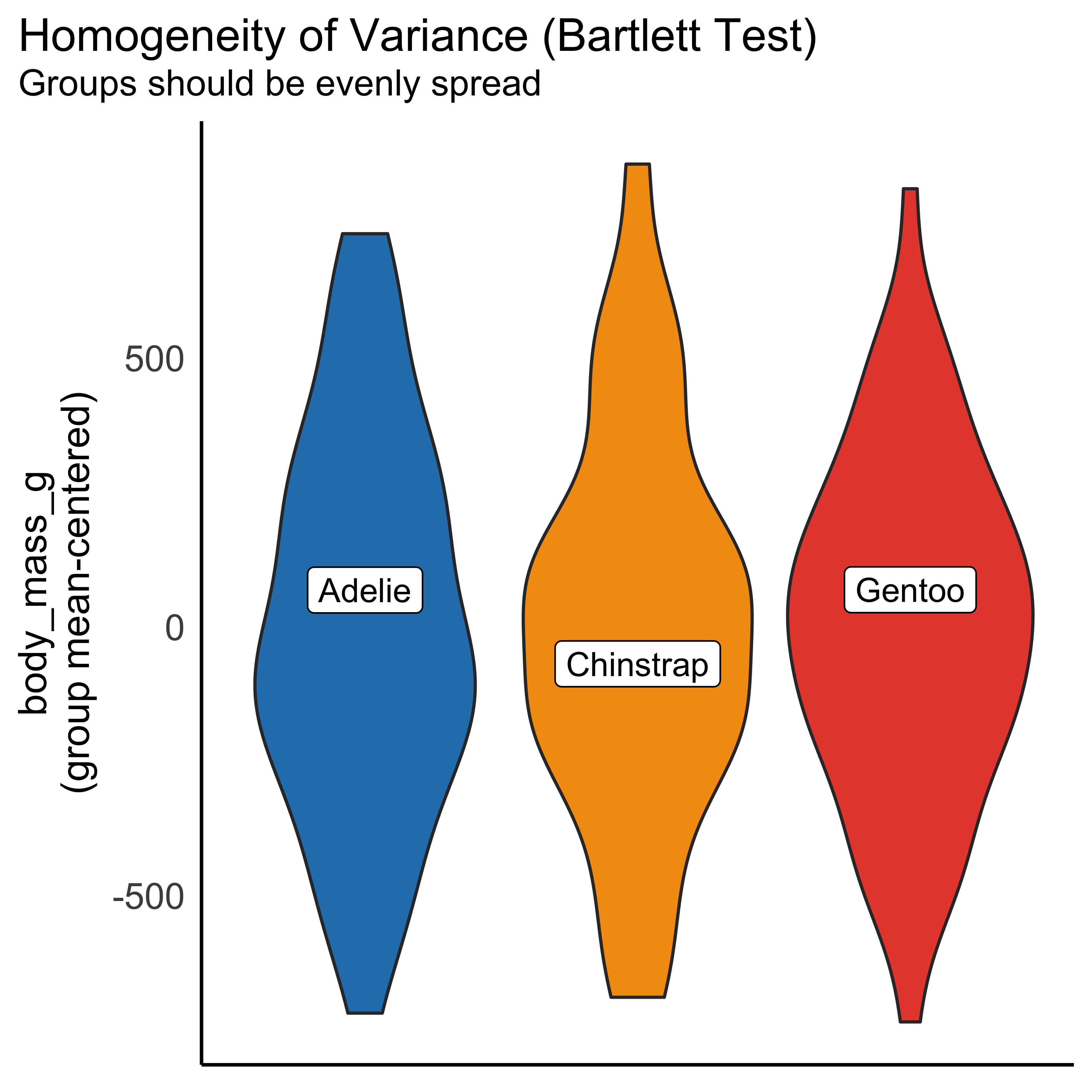
- Variance is homogenous/constant
- Residuals are normally distributed
Let’s try with an example
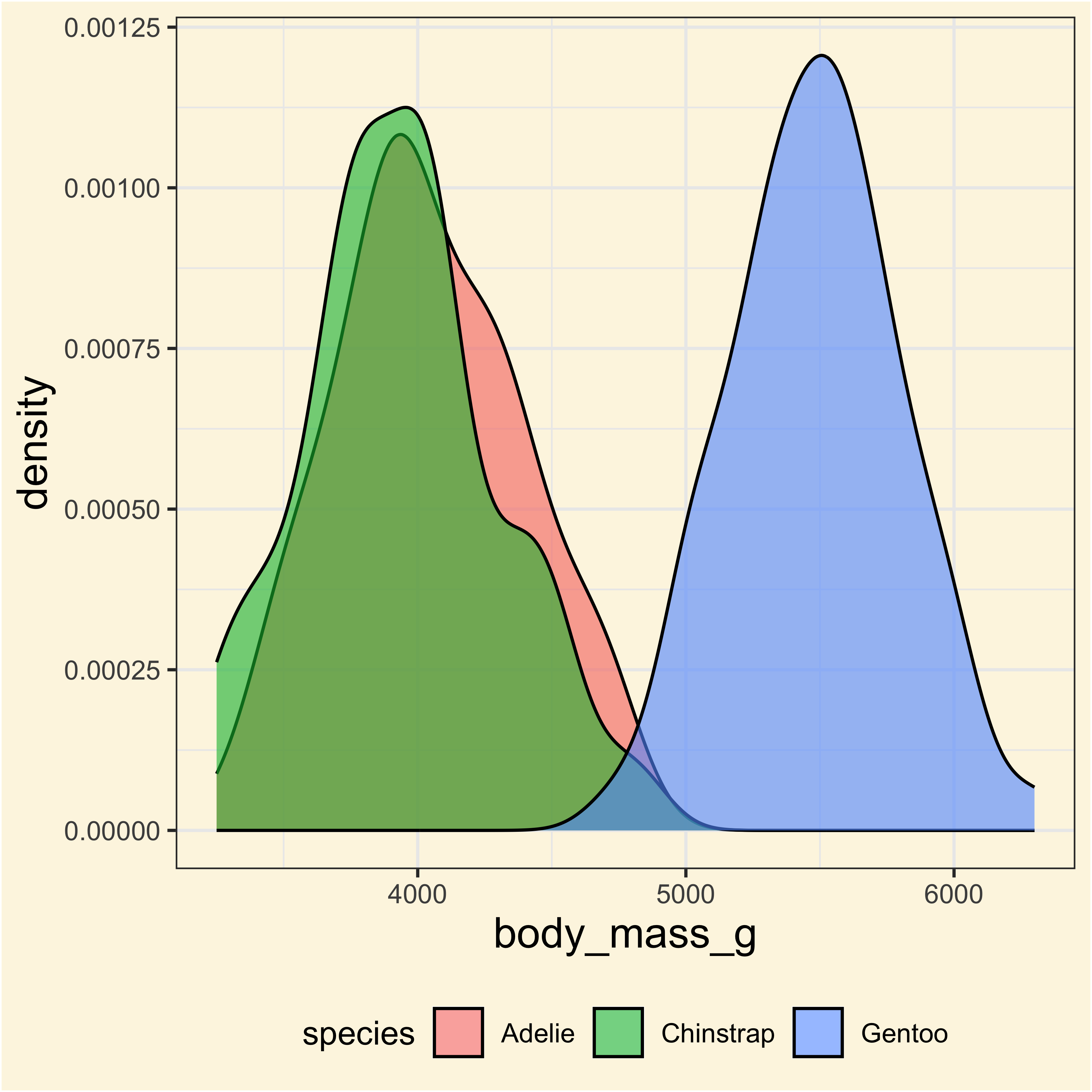
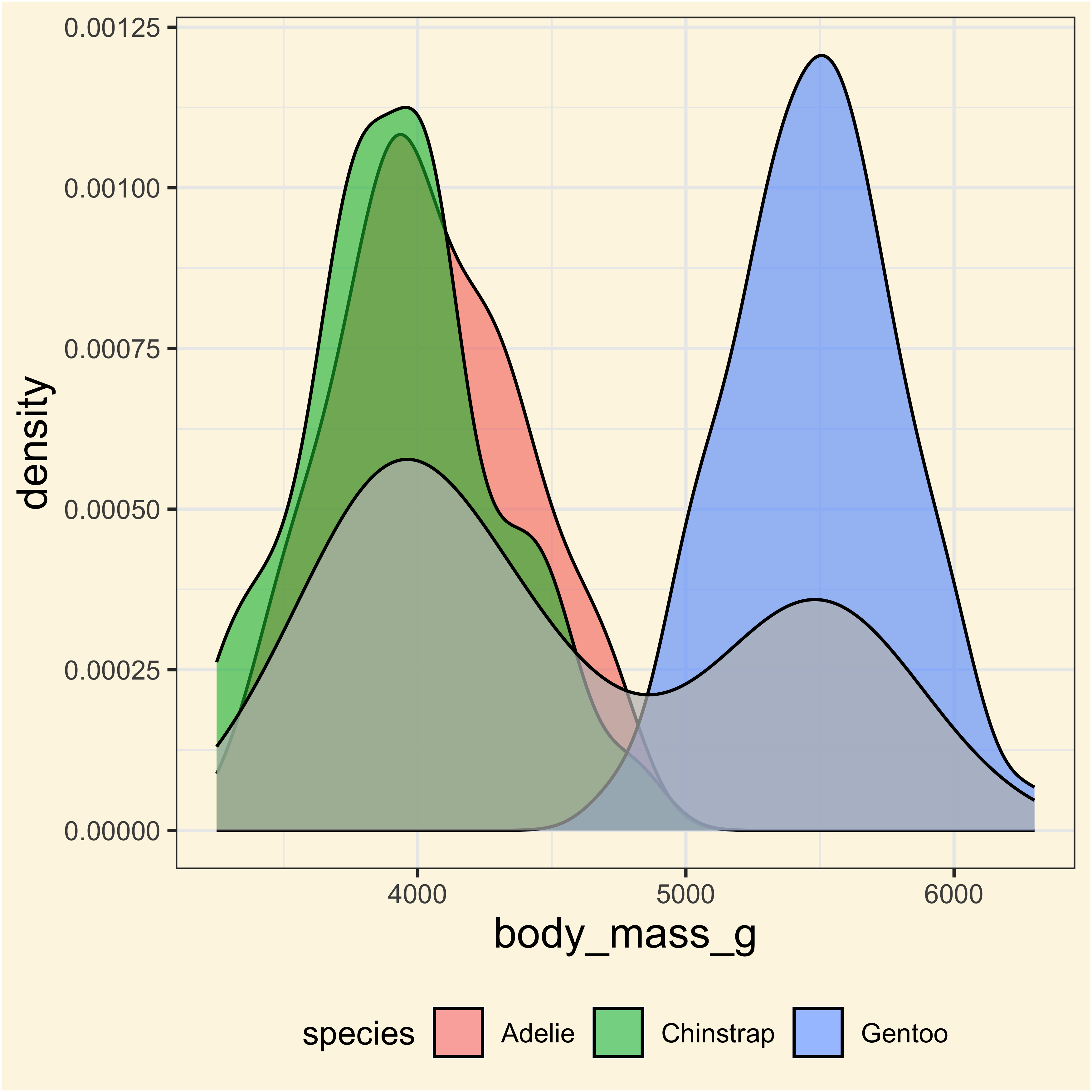
Applying ANOVA with different approaches
1. Using aov() function:
1. Using aov() function:
Df Sum Sq Mean Sq F value Pr(>F)
species 2 84728125 42364062 370.4 <2e-16 ***
Residuals 165 18871872 114375
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The Tukey-Krammer test to make pairwise comparisons:
TukeyHSD(anovamodel) Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = body_mass_g ~ species, data = males)
$species
diff lwr upr p adj
Chinstrap-Adelie -104.5226 -270.5952 61.55003 0.2990486
Gentoo-Adelie 1441.3429 1302.5929 1580.09292 0.0000000
Gentoo-Chinstrap 1545.8655 1374.6810 1717.04996 0.00000002. Using the statsExpression package:
anovatbl <- oneway_anova(data = penguins, x = species, y = body_mass_g)
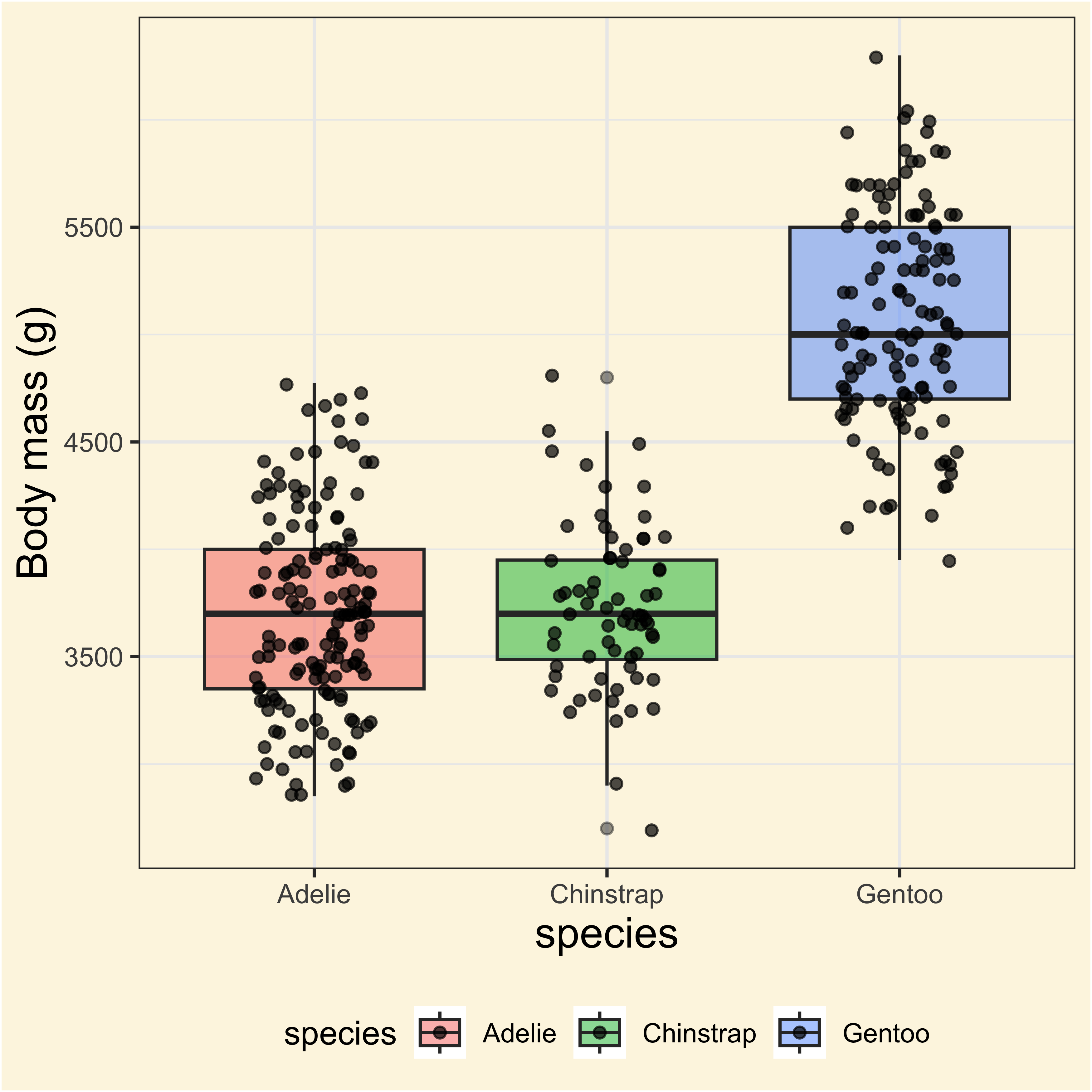
anovatblAdding stats to the plots!
ggplot(
males,
aes(
x = species,
y = body_mass_g,
fill = species
)
) +
geom_boxplot(alpha = 0.5) +
geom_jitter(width = 0.2, alpha = 0.7) +
labs(
y = "Body mass (g)",
subtitle = anovatbl$expression[[1]]
)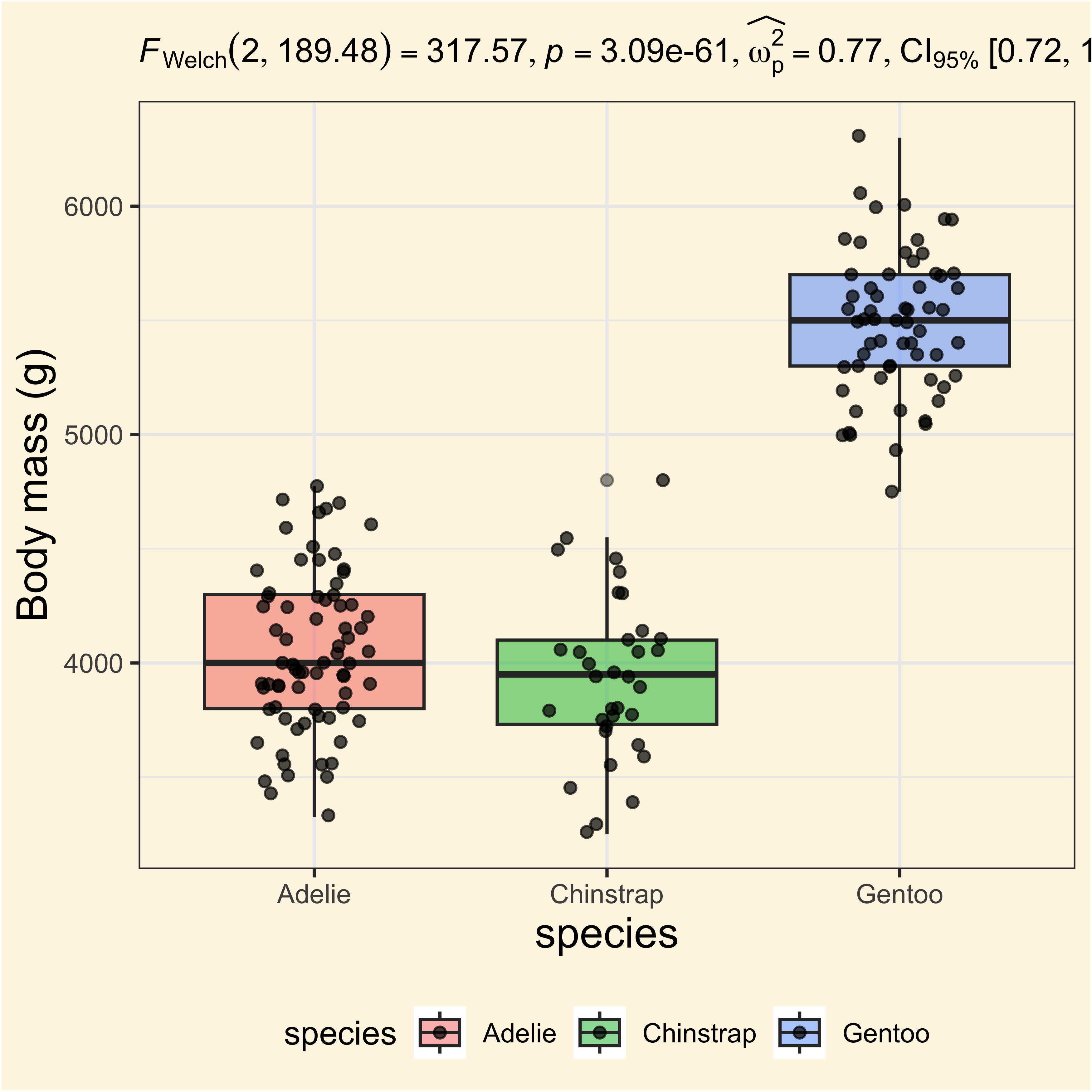
But why there are not comparisons?
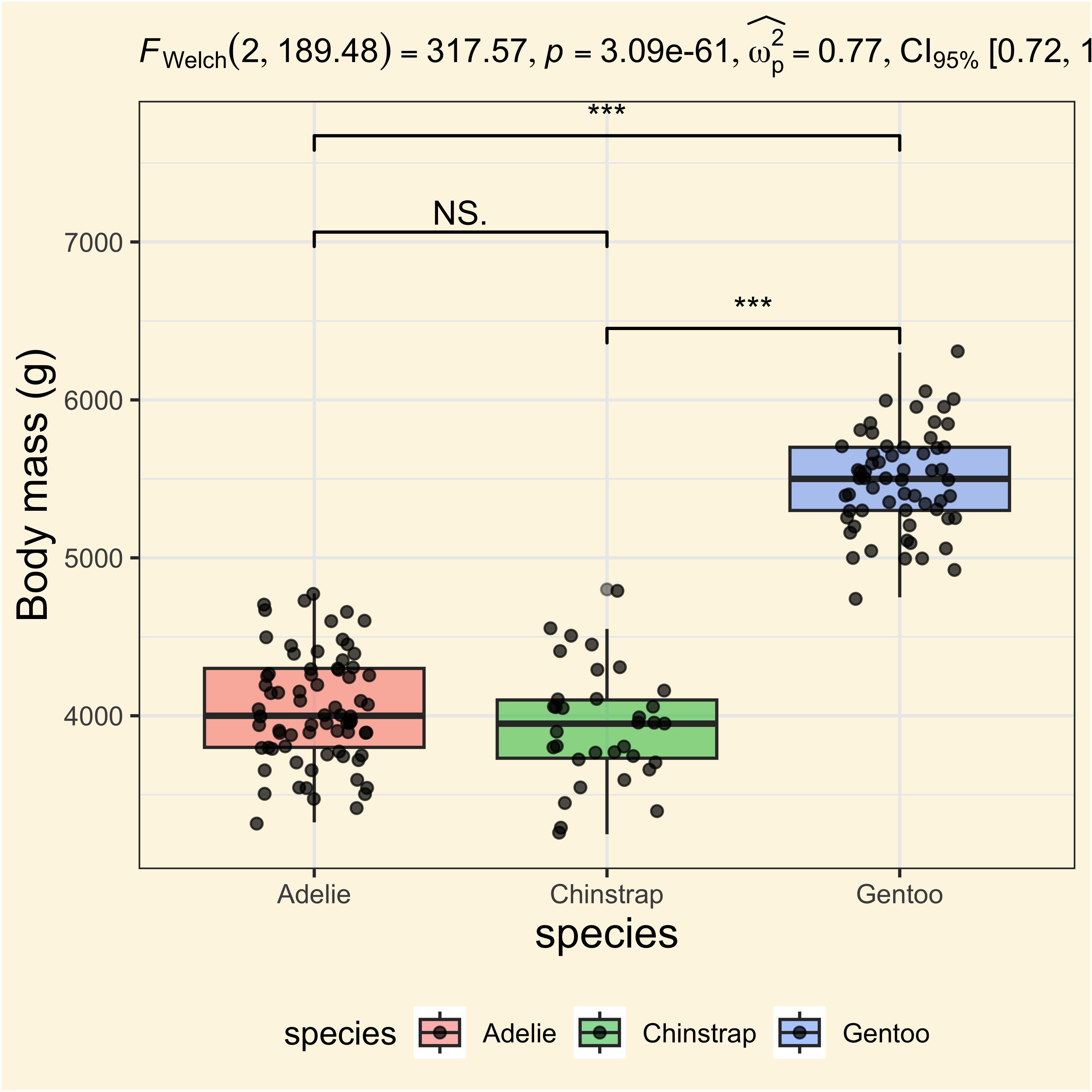
Adding pairwise comparisons
ggplot(
males,
aes(
x = species,
y = body_mass_g,
fill = species
)
) +
geom_boxplot(alpha = 0.5) +
geom_jitter(width = 0.2, alpha = 0.7) +
labs(
y = "Body mass (g)",
subtitle = anovatbl$expression[[1]]
) +
geom_signif(
comparisons = list(
c("Gentoo", "Chinstrap"),
c("Adelie", "Chinstrap"),
c("Adelie", "Gentoo")
),
step_increase = 0.2,
map_signif_level = TRUE
)
Testing the ANOVA assumptions with performance
- Sample and residuals independence:
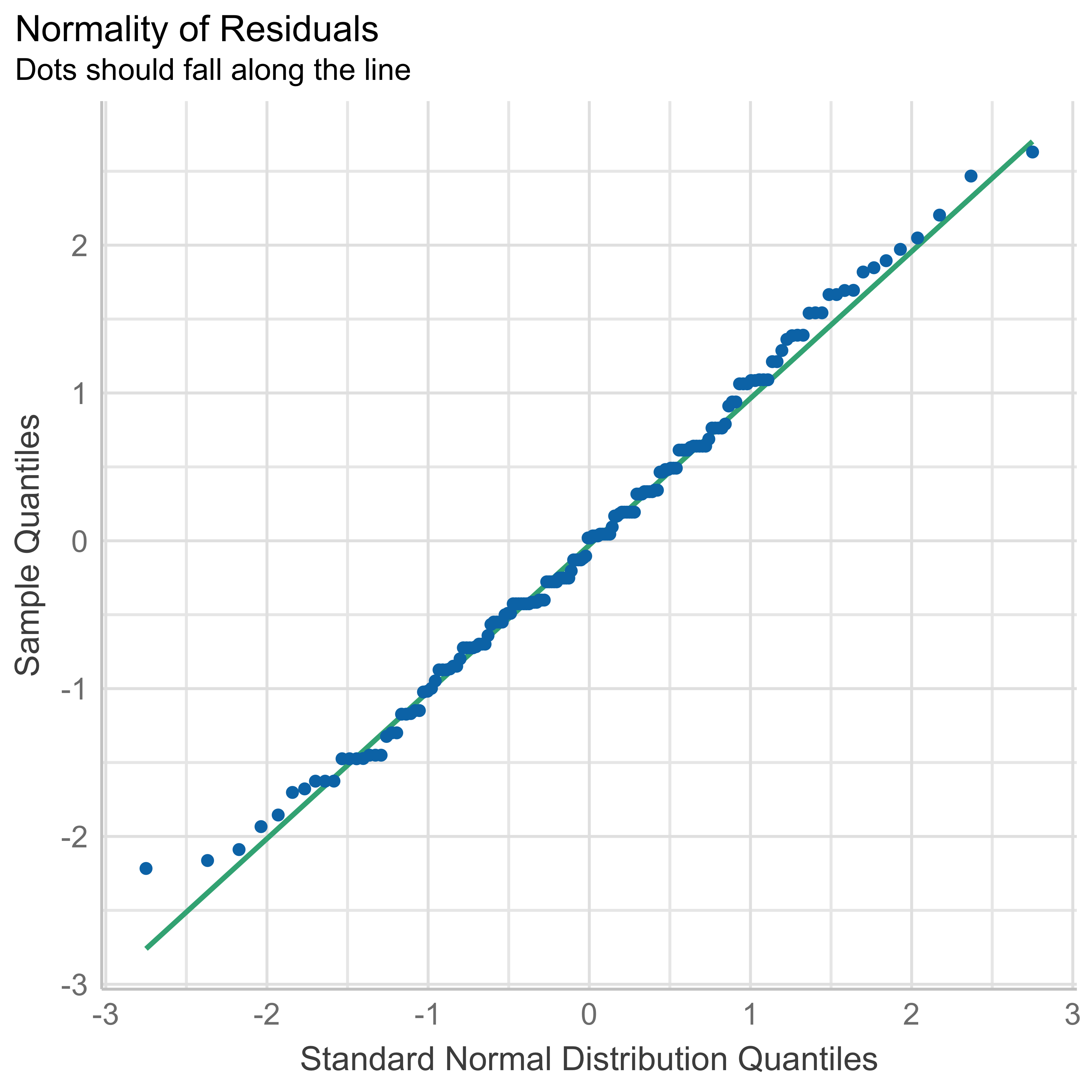
check_autocorrelation(anovamodel)OK: Residuals appear to be independent and not autocorrelated (p = 0.108).- Normality of the residuals:
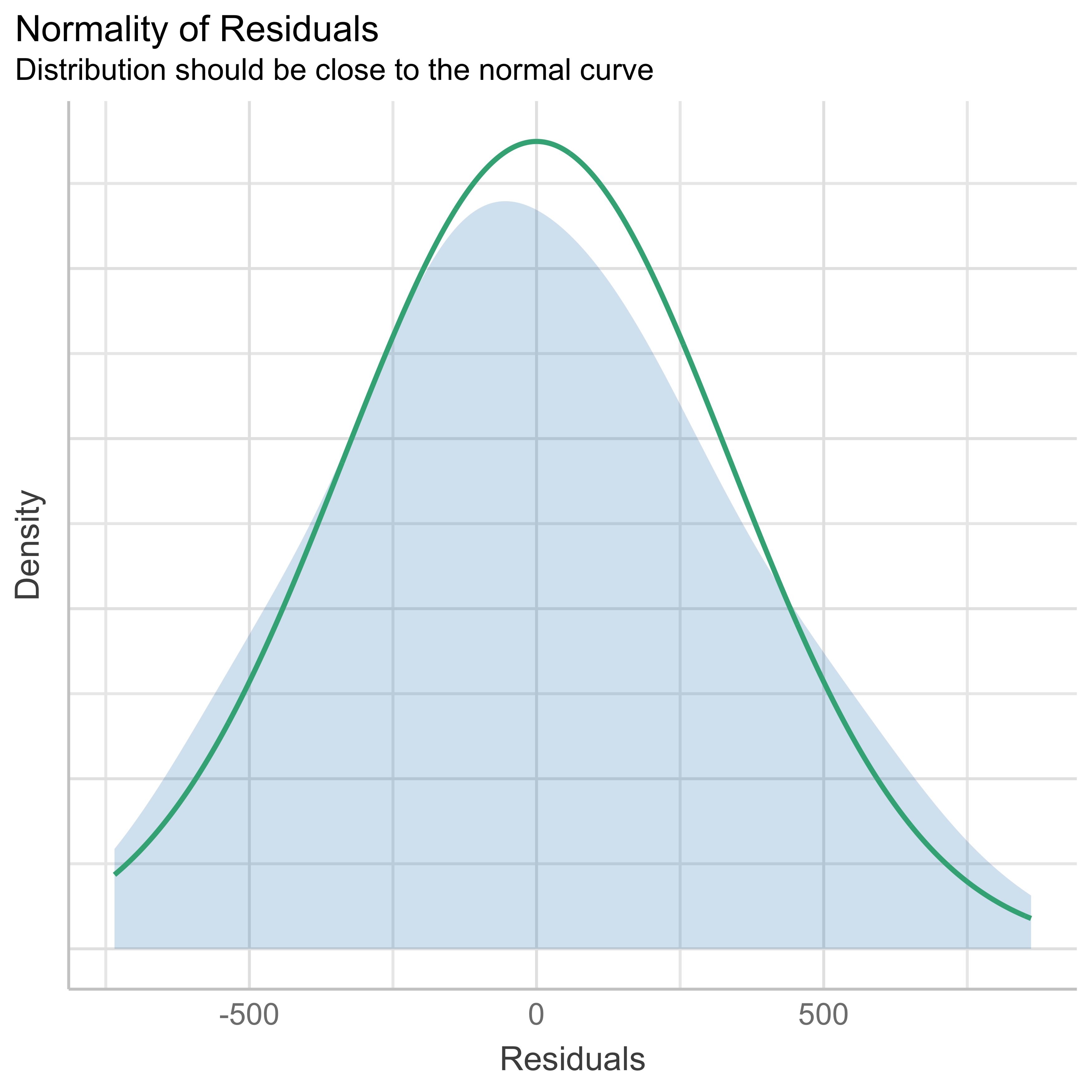
check_normality(anovamodel)OK: residuals appear as normally distributed (p = 0.484).- Homogeneity of the variance:
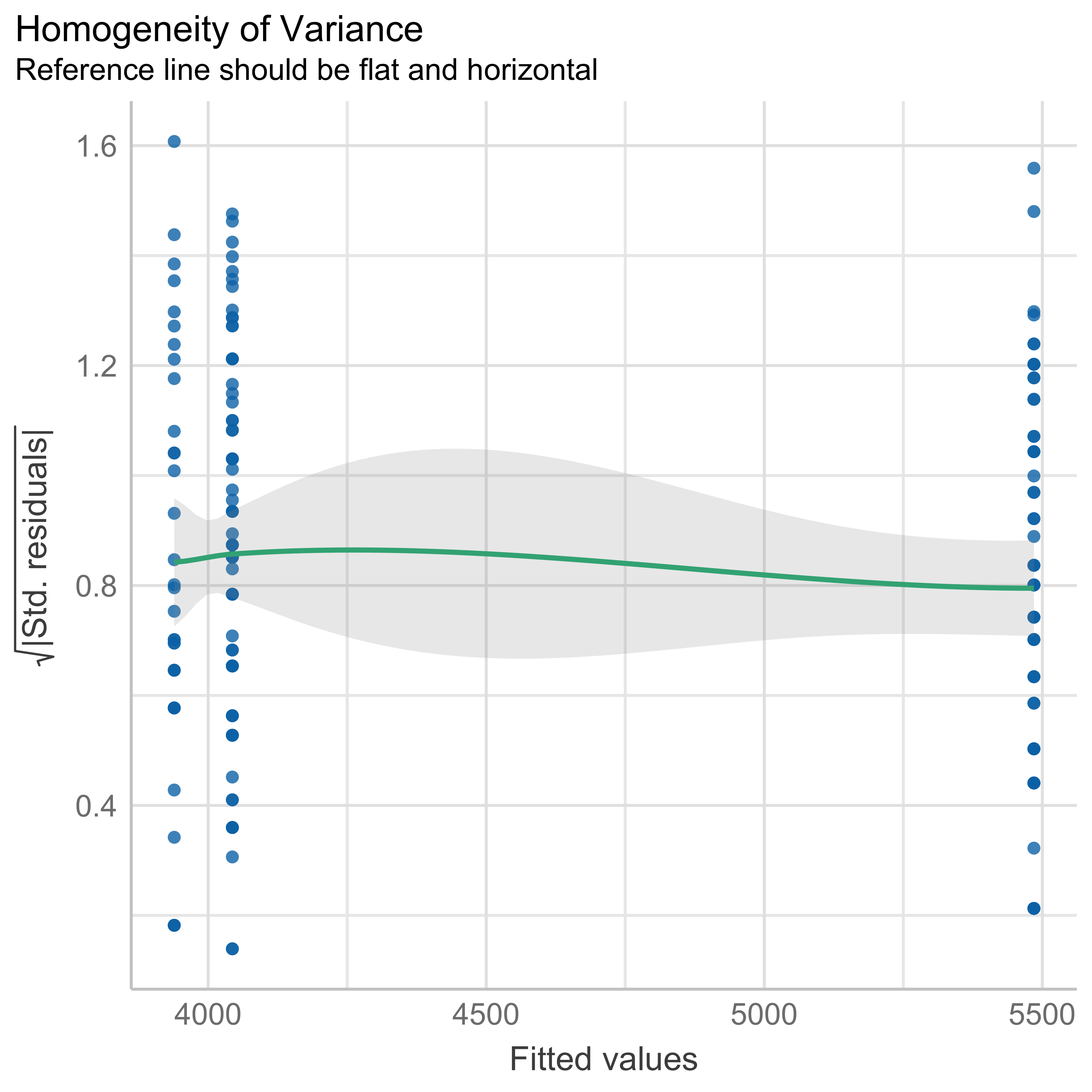
check_heteroskedasticity(anovamodel)OK: Error variance appears to be homoscedastic (p = 0.322).Checking influential observations (outliers)
check_outliers(anovamodel)OK: No outliers detected.
- Based on the following method and threshold: cook (0.5).
- For variable: (Whole model)What if normality and homogeneity of variance are not met then?
The Kruskal-Wallis test is a non-parametric alternative to the ANOVA test.
kruskal.test(data = males, body_mass_g ~ species)
Kruskal-Wallis rank sum test
data: body_mass_g by species
Kruskal-Wallis chi-squared = 116.5, df = 2, p-value < 2.2e-16BIOL2205 - Inferencia e Informática - DCB - Uniandes