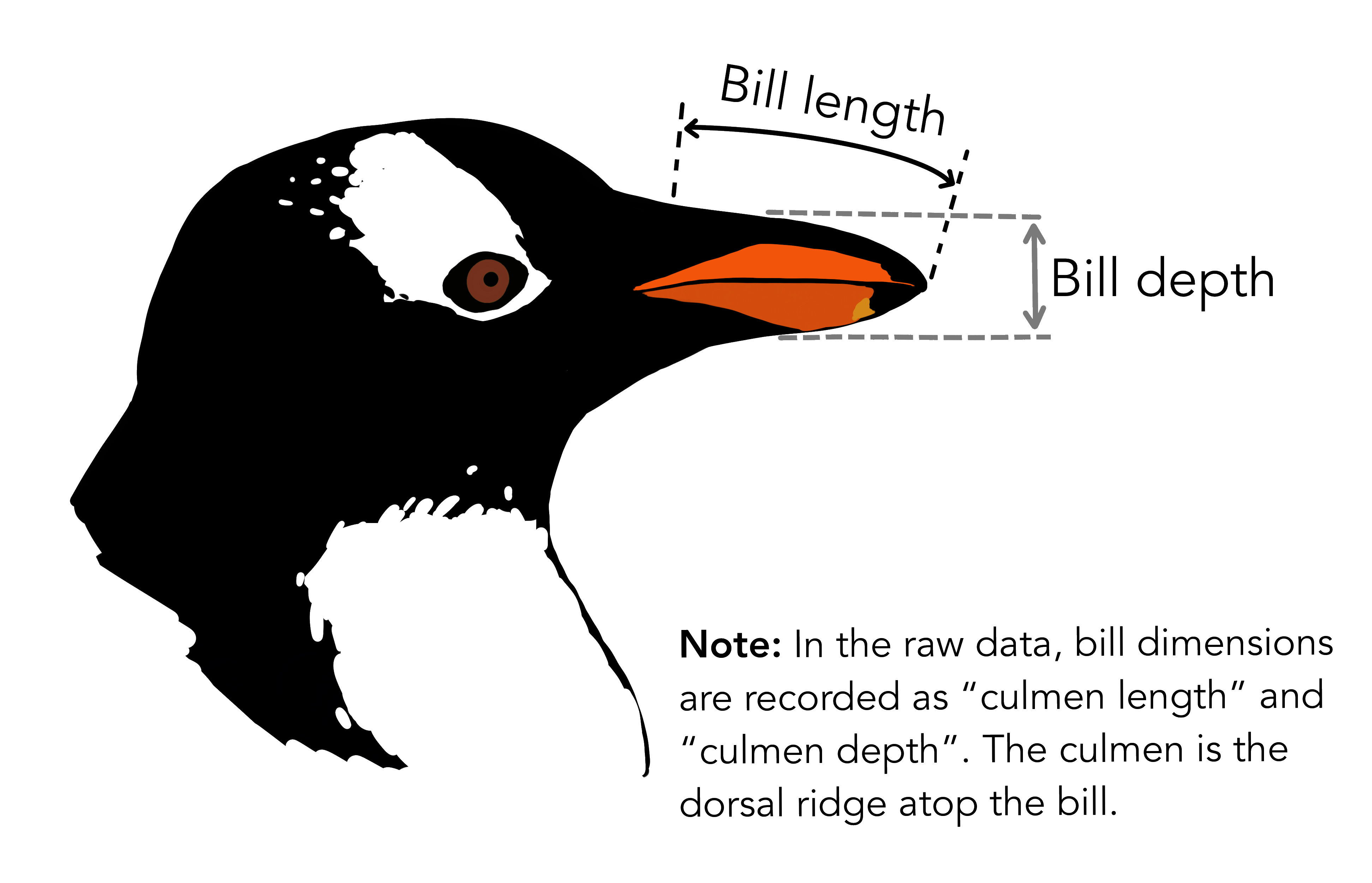
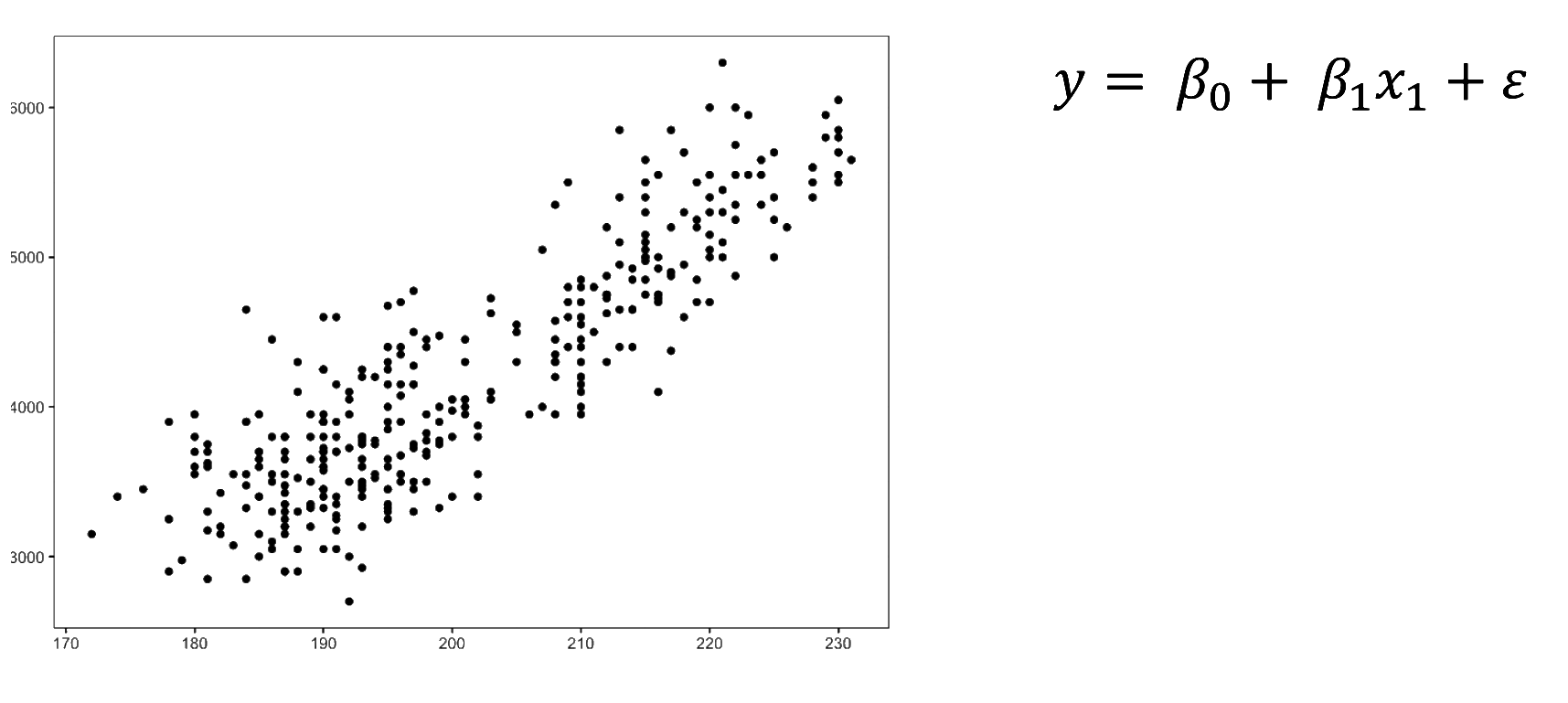
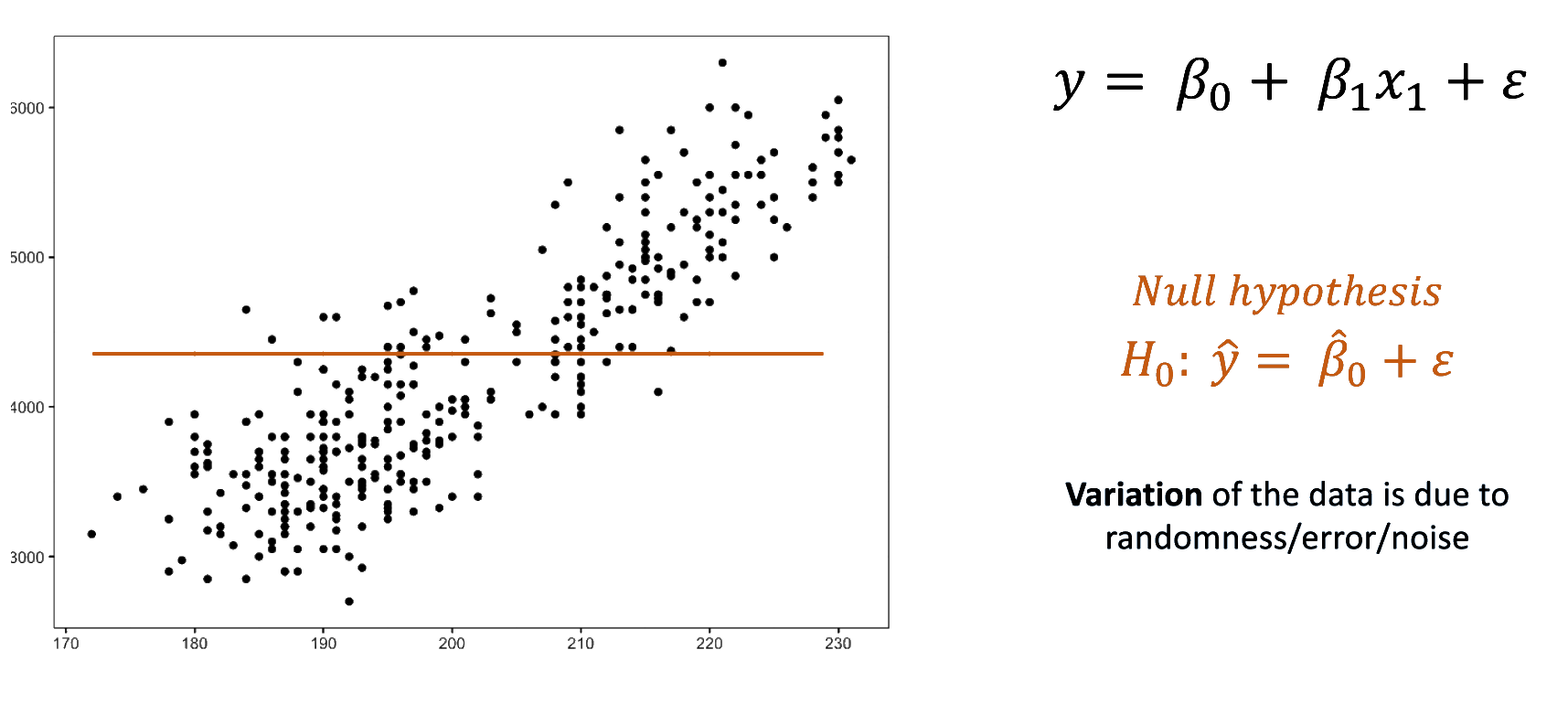
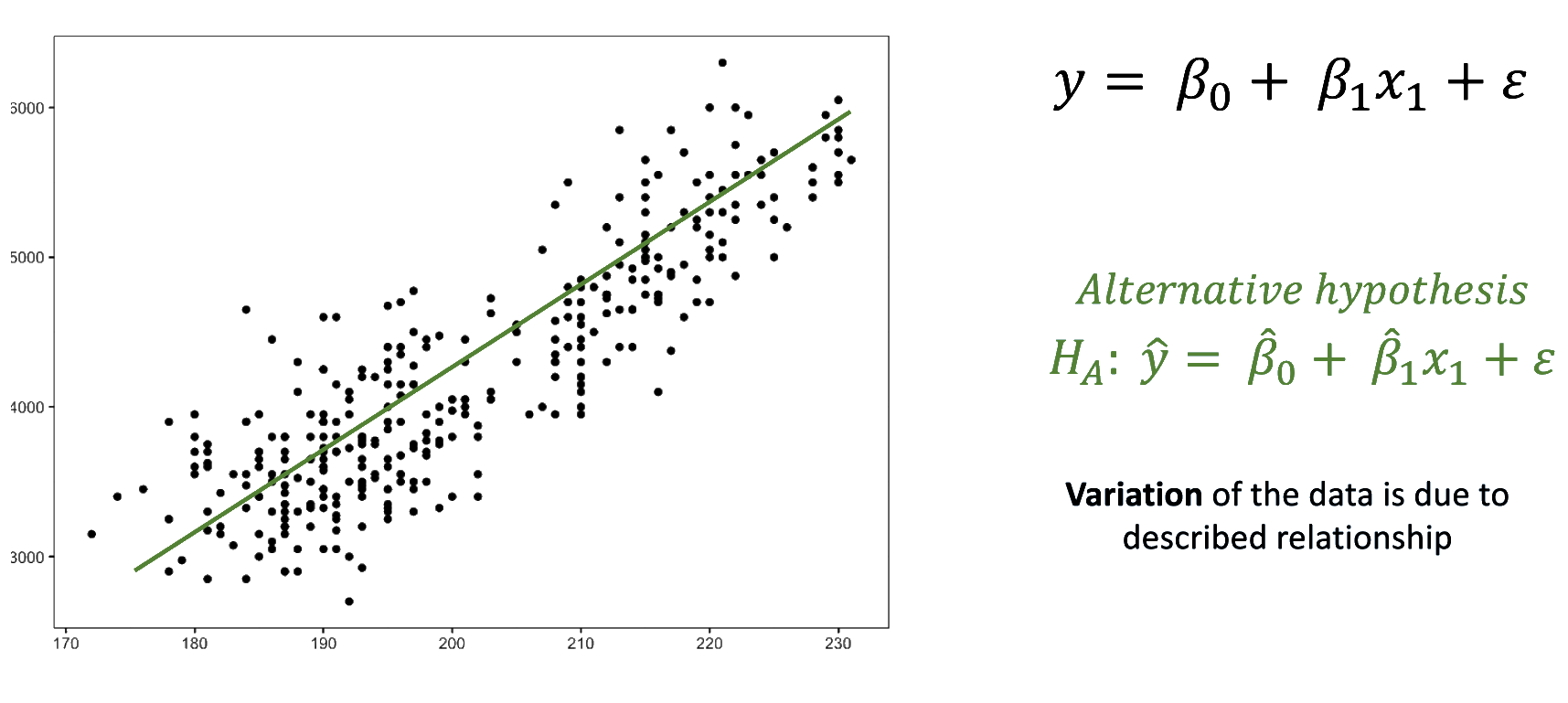
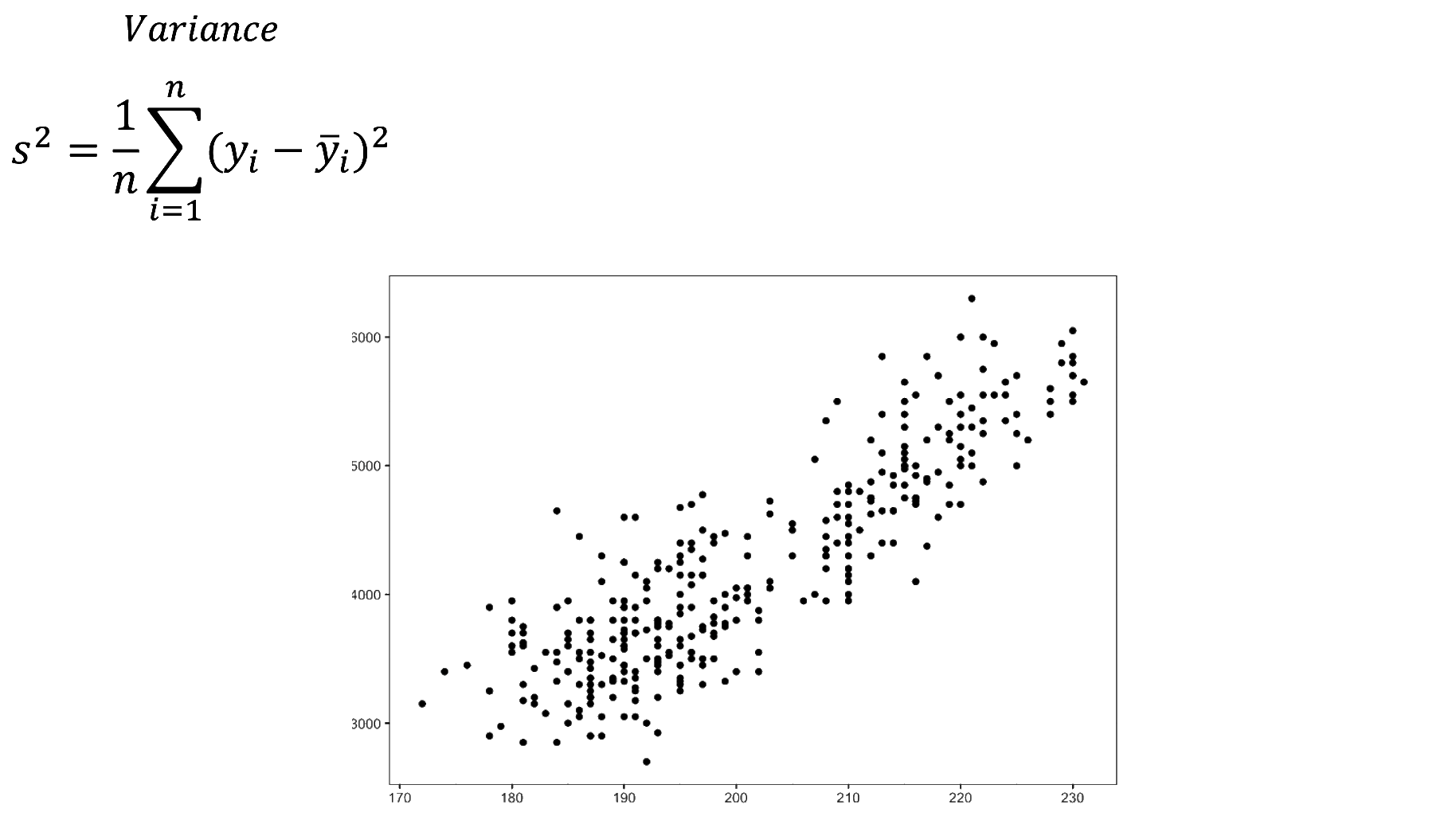
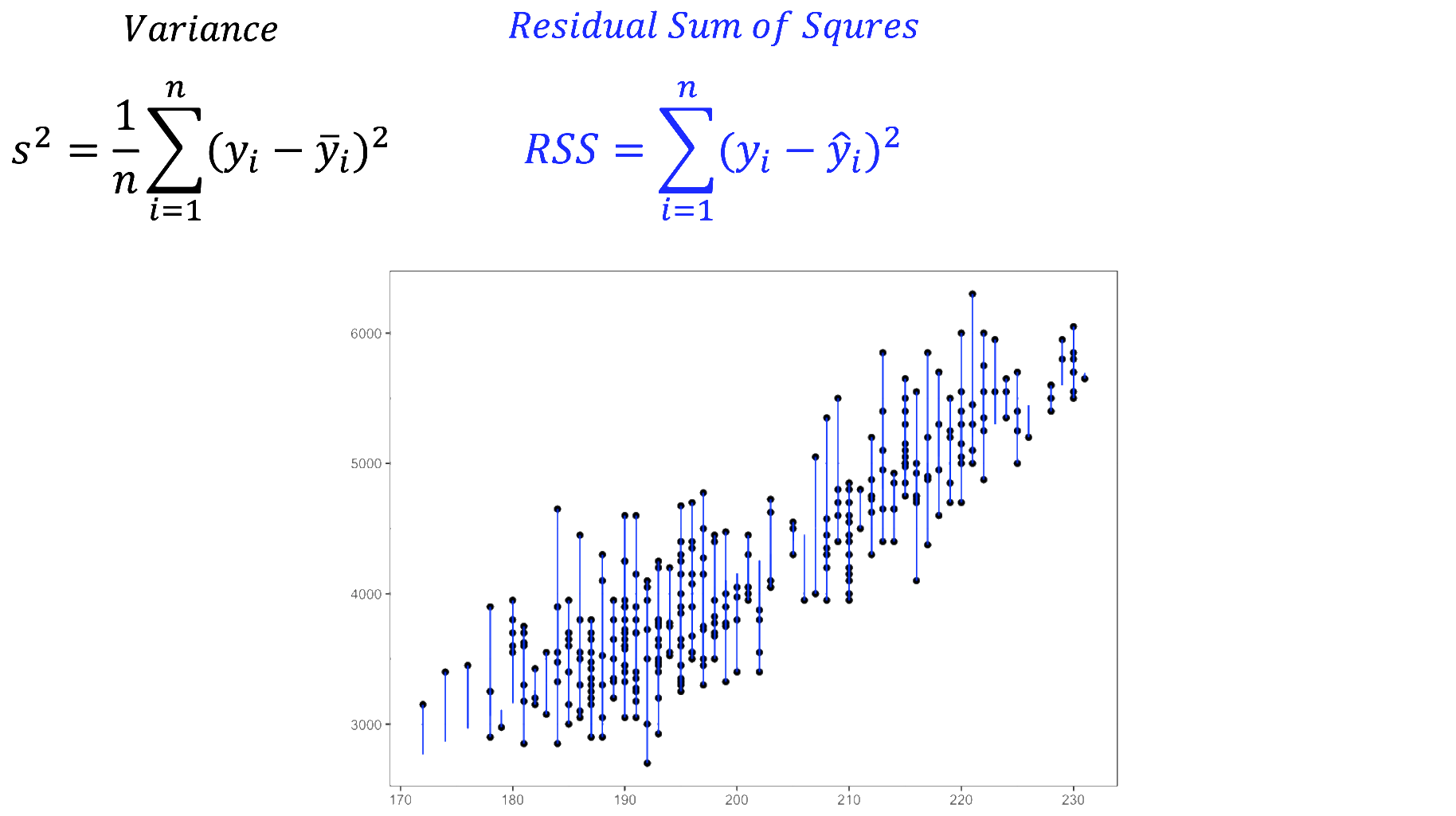
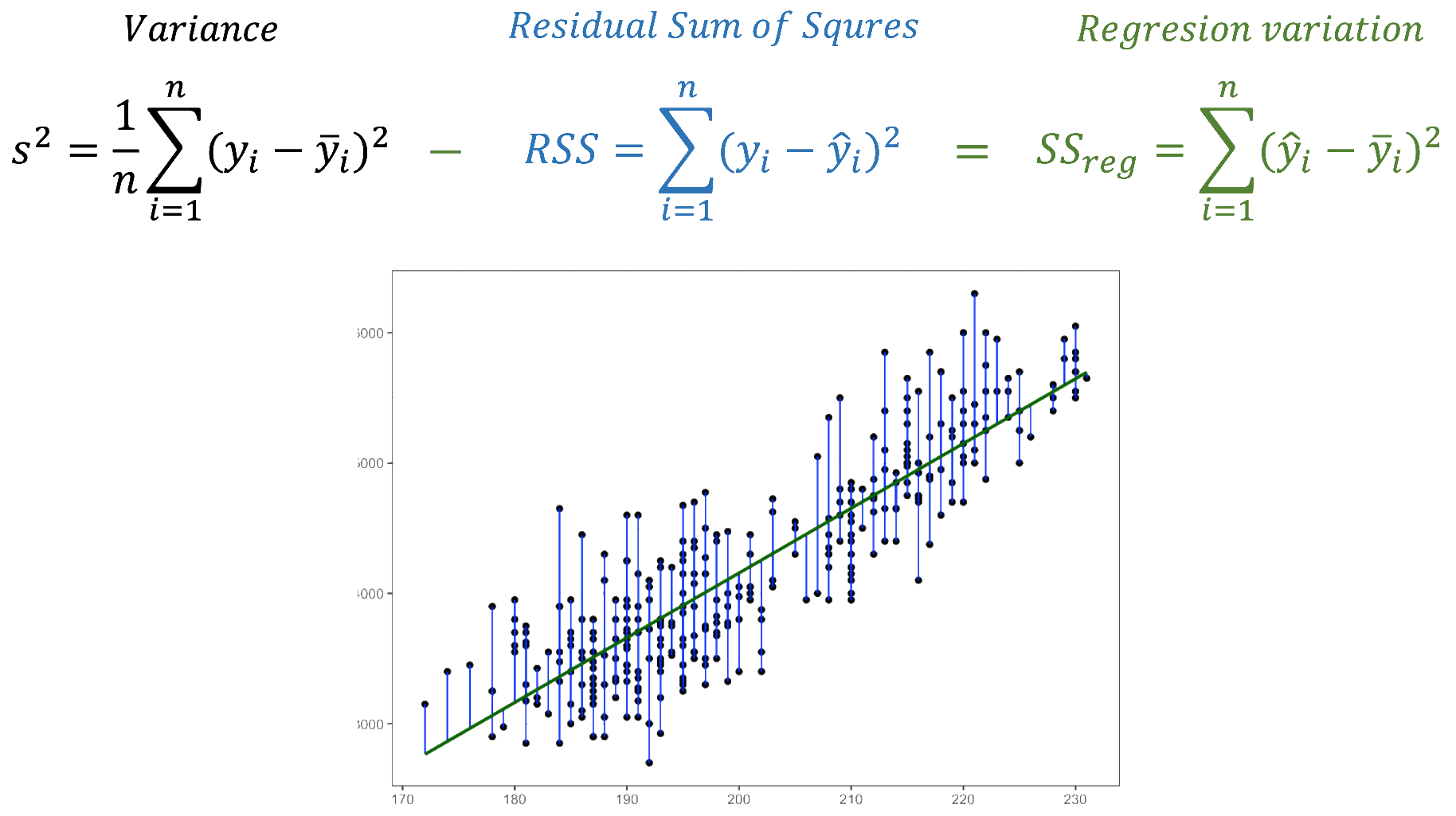
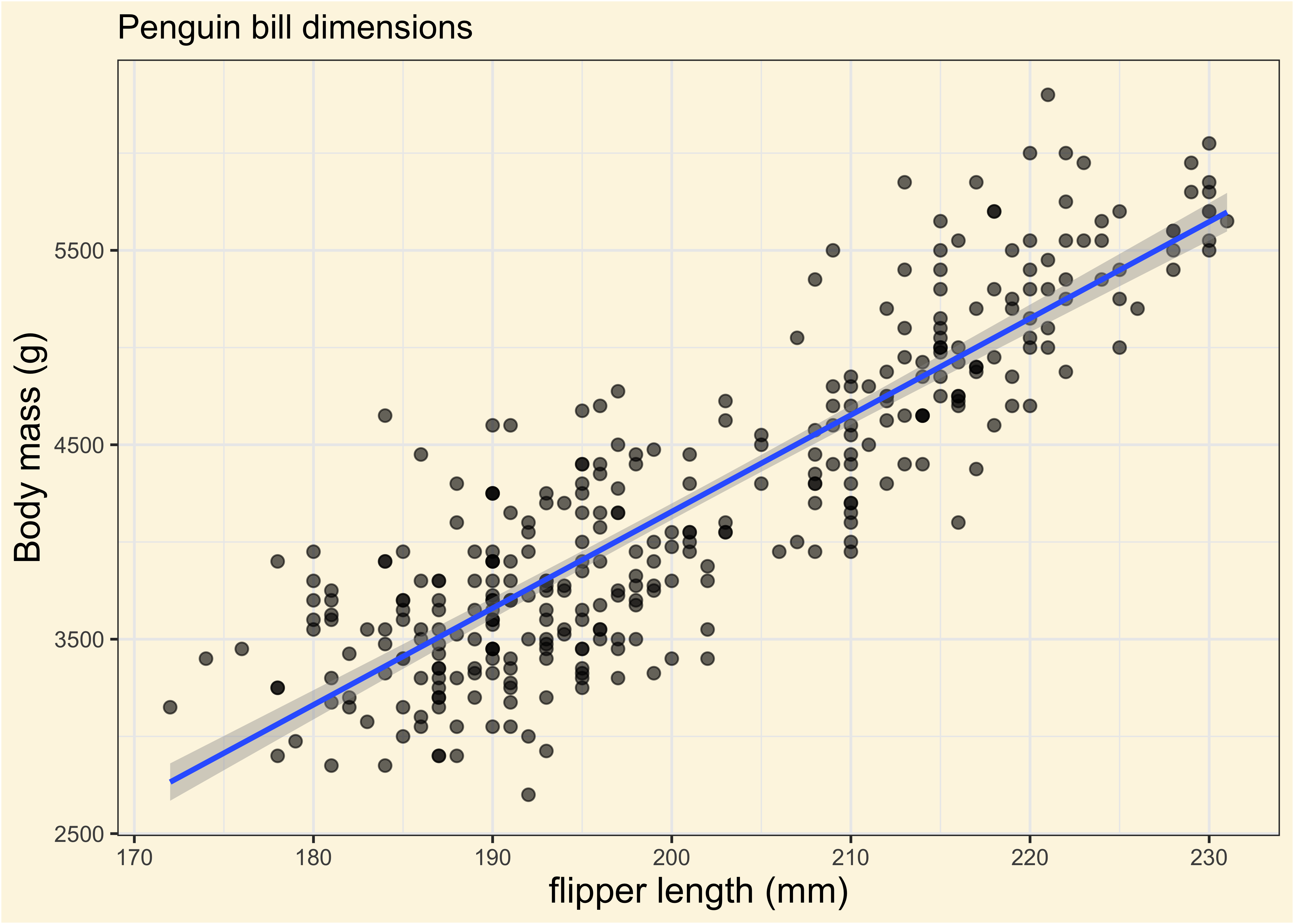
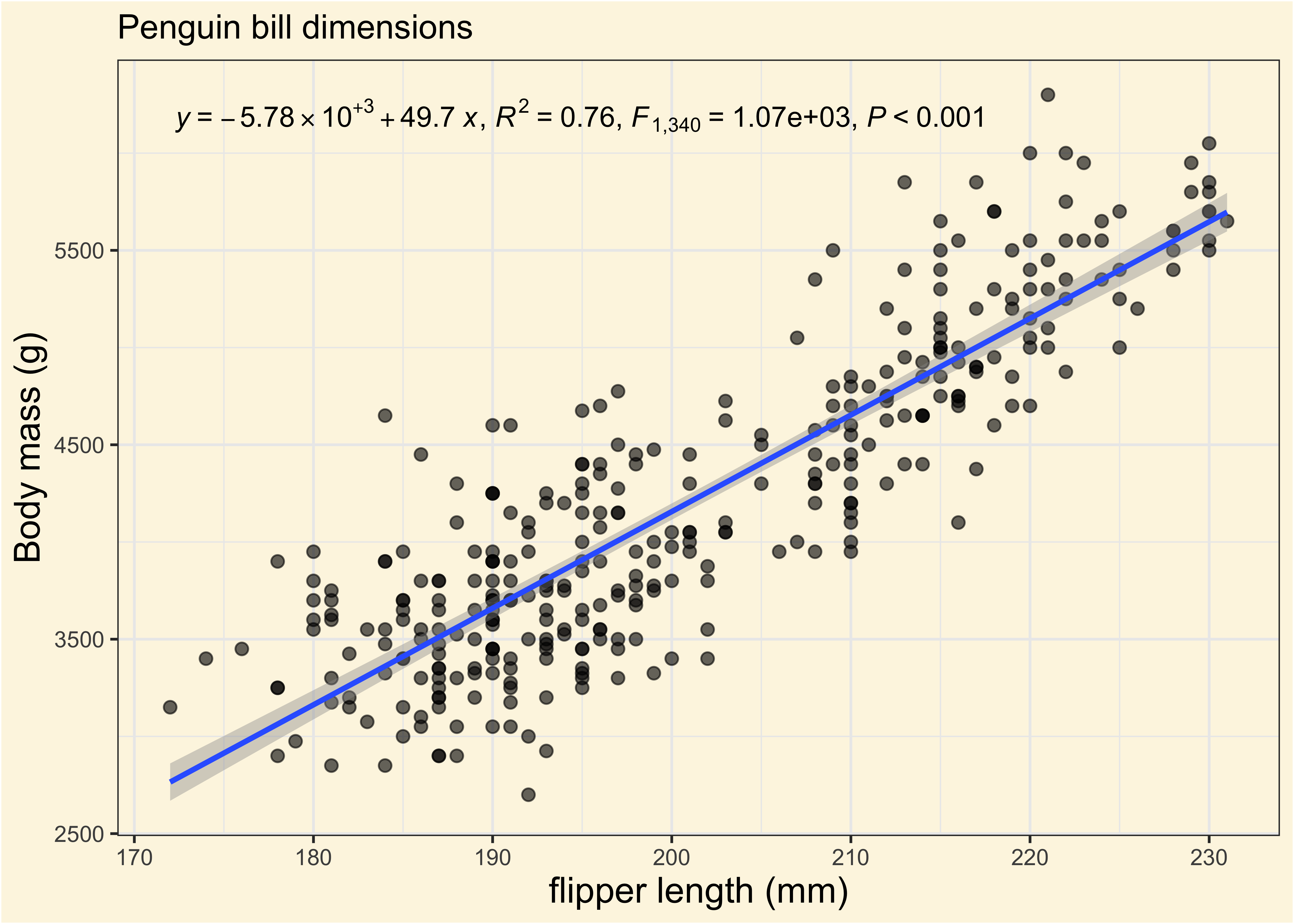
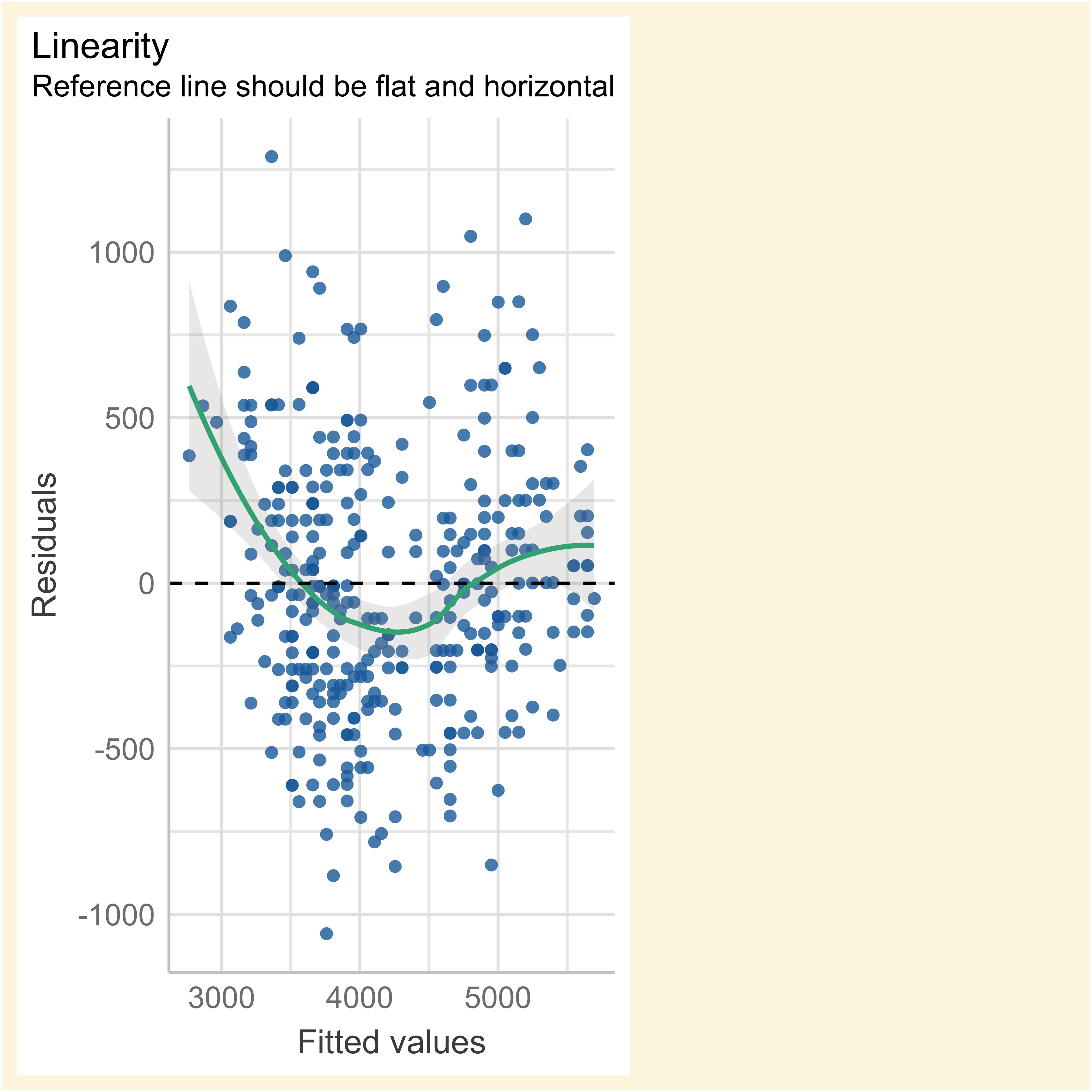
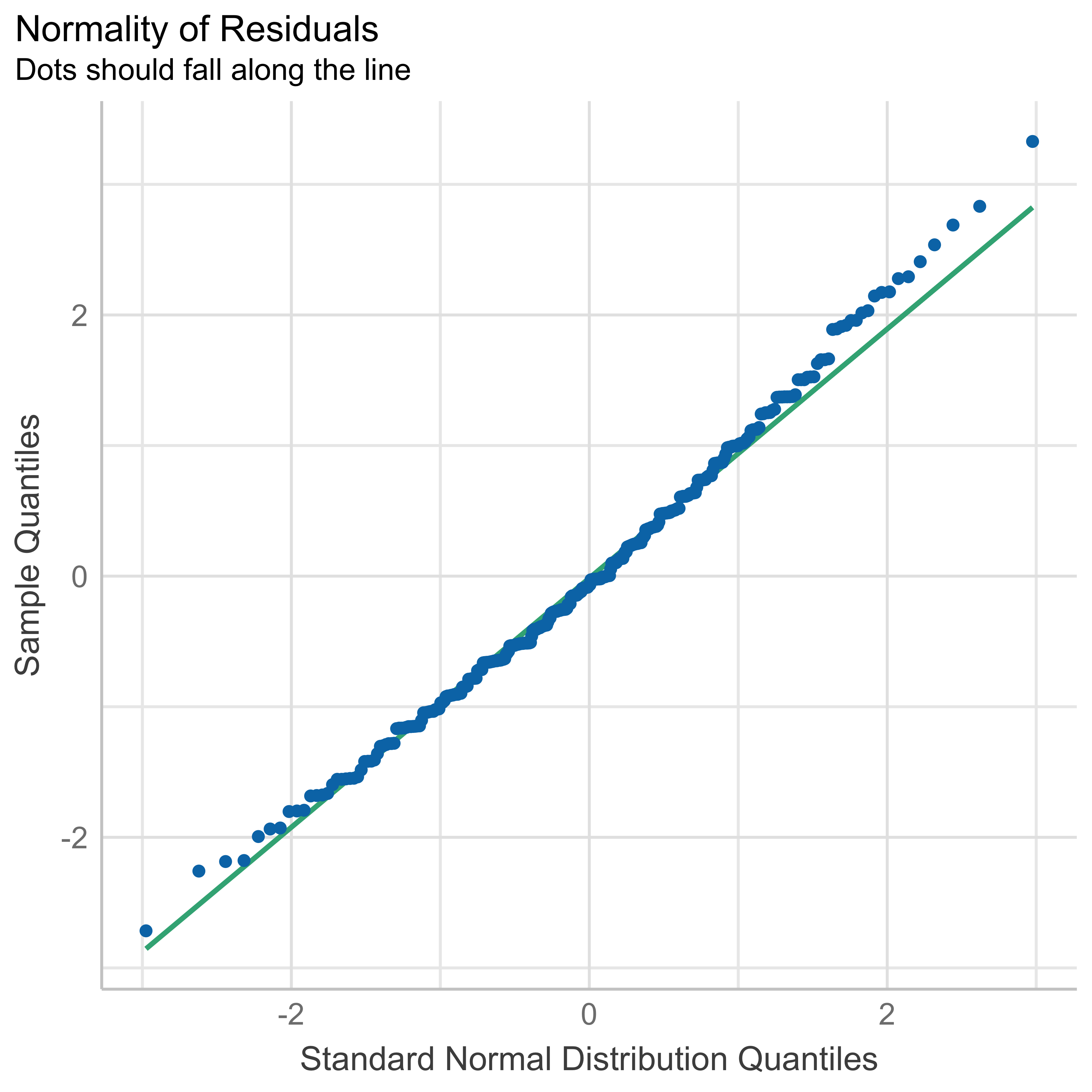
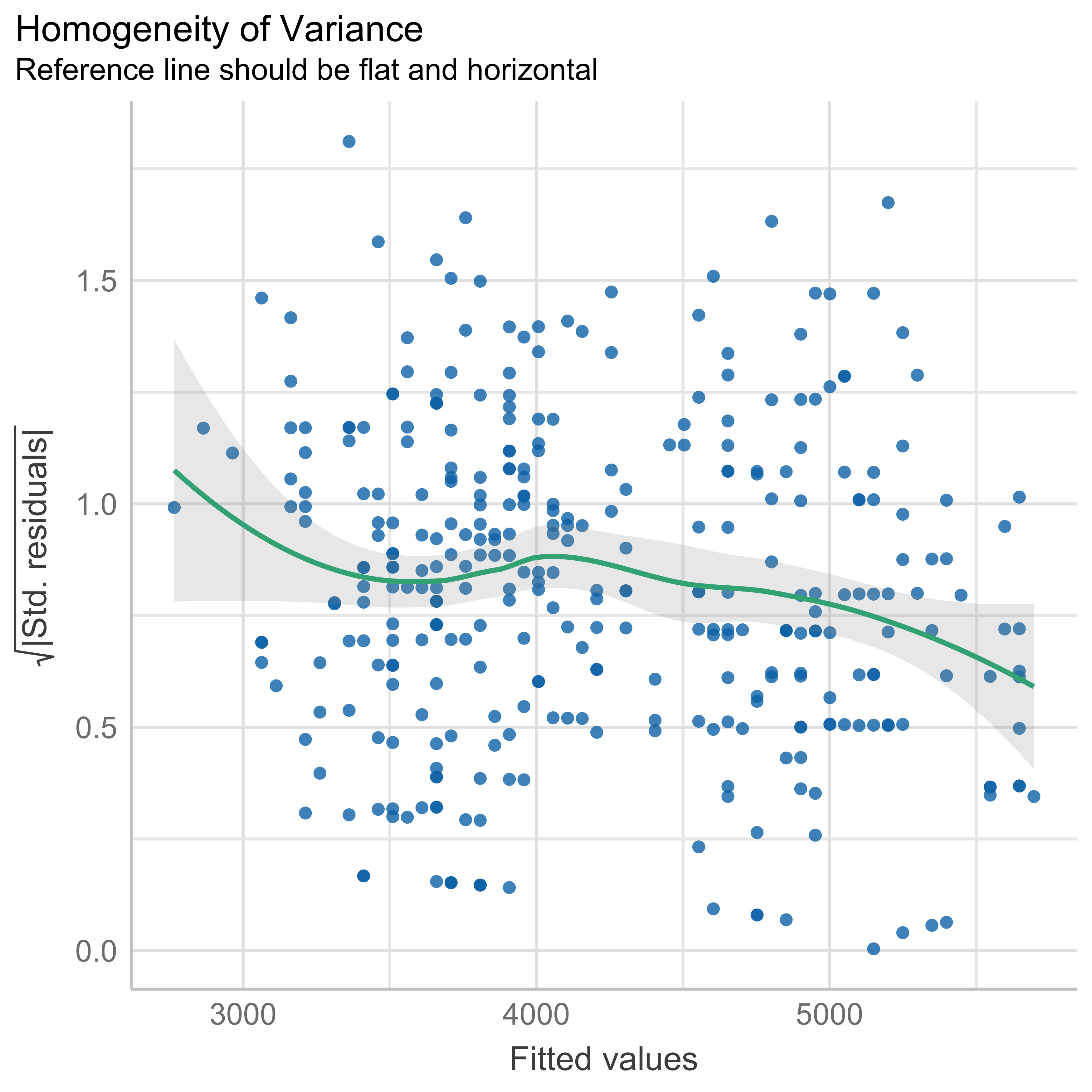
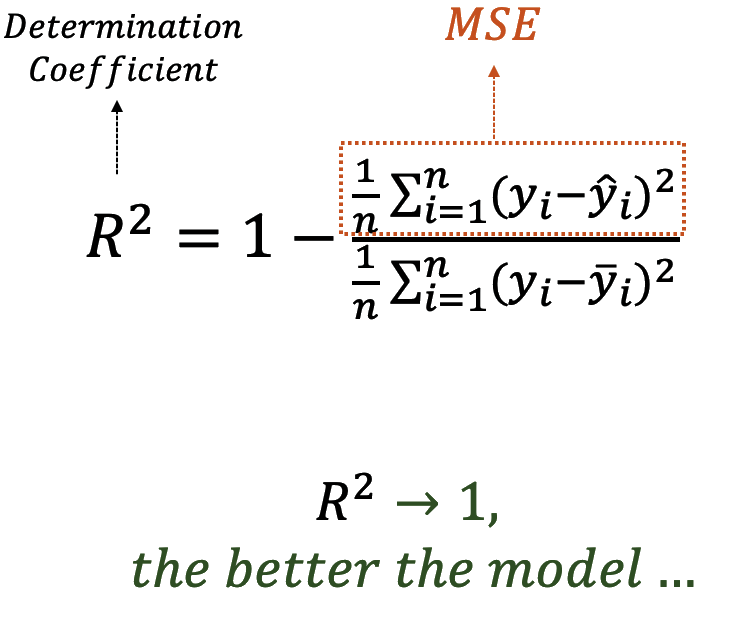We fitted a linear model (estimated using OLS) to predict body_mass_g with
flipper_length_mm (formula: body_mass_g ~ flipper_length_mm). The model
explains a statistically significant and substantial proportion of variance (R2
= 0.76, F(1, 340) = 1070.74, p < .001, adj. R2 = 0.76). The model's intercept,
corresponding to flipper_length_mm = 0, is at -5780.83 (95% CI [-6382.36,
-5179.30], t(340) = -18.90, p < .001). Within this model:
- The effect of flipper length mm is statistically significant and positive
(beta = 49.69, 95% CI [46.70, 52.67], t(340) = 32.72, p < .001; Std. beta =
0.87, 95% CI [0.82, 0.92])
Standardized parameters were obtained by fitting the model on a standardized
version of the dataset. 95% Confidence Intervals (CIs) and p-values were
computed using a Wald t-distribution approximation.