adelies <- penguins |>
filter(species == "Adelie") |>
drop_na() |>
mutate(binarysex = case_when(
sex == "male" ~ 1,
sex == "female" ~ 0
))GLM & AIC
https://tinyurl.com/29wkzwwj
Generalized Linear Models (GLM)
We have assume data (and residuals) follow a normal distribution for regular lineal models, but what if that does not always happen?
Structural component (\(\eta = \beta x\))
Link function (\(\eta = g(\mu)\))1
Random component (Normal, Binomial, Poisson, Neg. Binomial, etc.)
What regression should we apply?
It depends on the outcome datatype and on the errors distribution of the model.
| Regression | Outcome variable | Errors distribution | Link function |
|---|---|---|---|
| Regular/Estandar | Continuos | Normal | Indentity |
| Logistic | Discrete (two-level factor - binary) | Binomial | Logit |
| Poisson | Skewed discrete (counts) | Poisson | Log |
| Neg. Binomial | Discrete (counts) | Neg. Binomial | Inverse |
| Gamma | Skewed positive continuous | Gamma | Reciprocal |
Let’s try it in R:
Let’s create the GLM for to predict a penguin’s sex based on it’s body mass:
binarysexmodel <- glm(binarysex ~ body_mass_g, data = adelies, family = "binomial")Let’s see what the outout is:
Call: glm(formula = binarysex ~ body_mass_g, family = "binomial", data = adelies)
Coefficients:
(Intercept) body_mass_g
-28.749639 0.007814
Degrees of Freedom: 145 Total (i.e. Null); 144 Residual
Null Deviance: 202.4
Residual Deviance: 88.74 AIC: 92.74What did we do?
report::report(binarysexmodel)We fitted a logistic model (estimated using ML) to predict binarysex with
body_mass_g (formula: binarysex ~ body_mass_g). The model's explanatory power
is substantial (Tjur's R2 = 0.61). The model's intercept, corresponding to
body_mass_g = 0, is at -28.75 (95% CI [-39.49, -20.42], p < .001). Within this
model:
- The effect of body mass g is statistically significant and positive (beta =
7.81e-03, 95% CI [5.55e-03, 0.01], p < .001; Std. beta = 3.58, 95% CI [2.54,
4.92])
Standardized parameters were obtained by fitting the model on a standardized
version of the dataset. 95% Confidence Intervals (CIs) and p-values were
computed using a Wald z-distribution approximation.Let’s put that on a plot
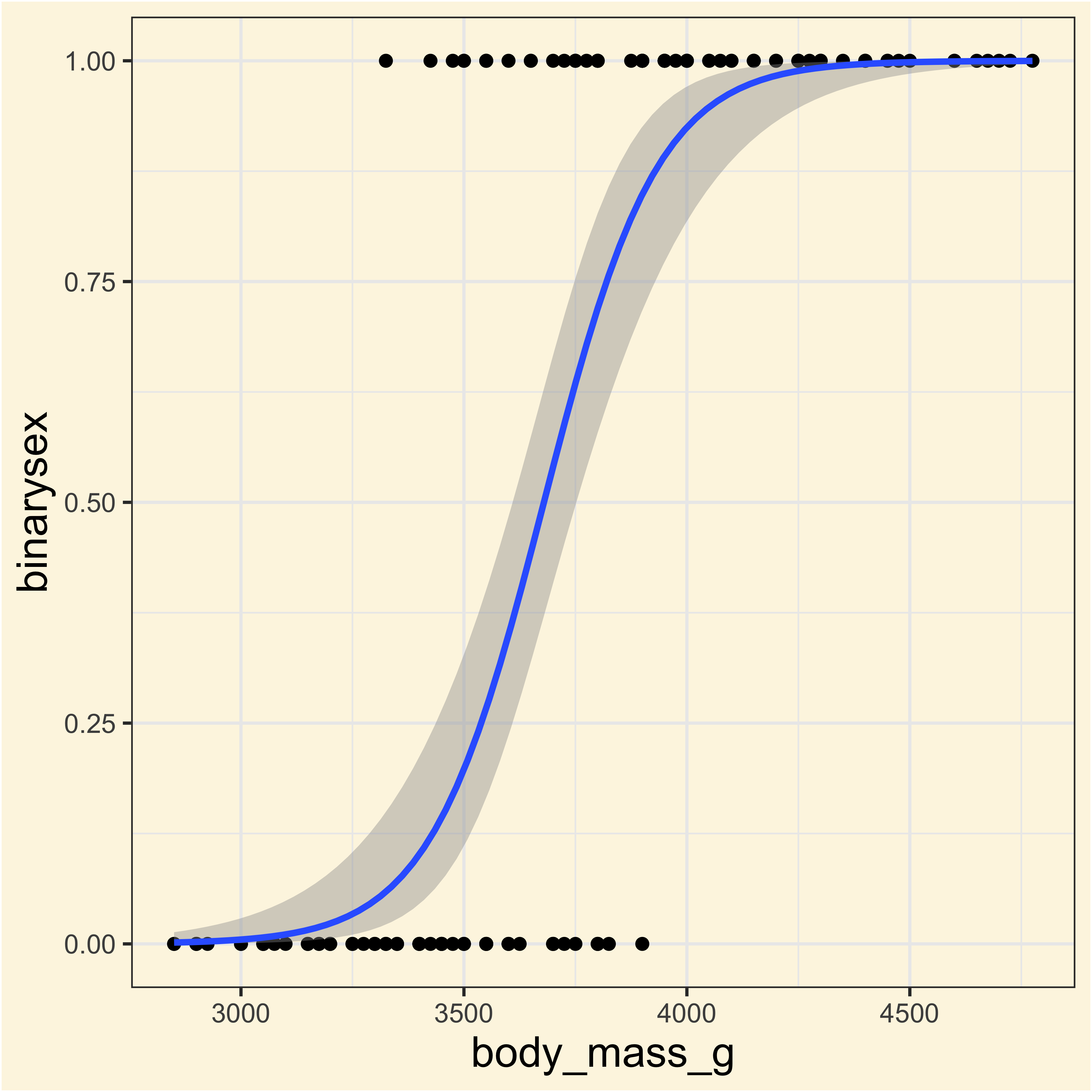
ggplot(
adelies,
aes(x = body_mass_g, y = binarysex)
) +
geom_point() +
geom_smooth(
method = "glm",
method.args = list(
family = "binomial"
)
)
Akaike Information Criteria (AIC)
An information theory concept that evaluates mathematical models in terms of its complexity and performance (prediction):
\[AIC=−2*ln(L)+2*k\]
Using Broom to evaluate statistical models.
Tidying model’s statistical information and report their performance.
tidy()Summarizes and organize the model informationaugment()Displays all model information for each data point.glance()Summarizes the model performance (AIC, BIC, and others).
broom::tidy(binarysexmodel)broom::augment(binarysexmodel)Now let’s see the AIC:
broom::glance(binarysexmodel)BIOL2205 - Inferencia e Informática - DCB - Uniandes