Introduction
The present book gathers the lecture notes of the undergrad course in Fundamentals of Computational Biology that takes place in EAFIT University. The first part will focus on how to use the the command line interface (aka CLI). It includes a long-format chapter about the Unix tools and concepts that is taught during the first four class lessons. The second chapter of covers the basics of git and the principal workflows to work daily on a collaborative manner this is actually the second lesson of the course. The third lessons, highlights the importance of package manager systems (such as homebrew and conda or scoop) and briefly introduce the main concepts and relevant commands. Later, in a fourth lesson, we introduce important concepts about how computers handle the tasks or jobs, and the parallelization of them. We talk about the computer architectures, the cluster architectures and the job scheduling software called Slurm, which is used on our local HPC cluster.
The second part is dedicated to sequence analysis and will cover several topics related to sequencing technologies, sequence alignments. All this topics are covered from the biological perspective, mathematical notions and the practical computing approach. Some of the main bioinformatics file extensions are covered and the git
Third part is focused genomics. It will cover main algorithms for genome assembly and some relevant programs. Also the biological nuances of gene calling and gene annotation, and finally will variant calling analysis, which introduces the gene mapping concept and some of the most important bioinformatics file extensions.
Next parts are currently optionals for the course per-se and some iterations may only include one of those or some combinations. They are mainly dedicated to introduce metagenomics, differential gene expression analysis and structural bioinformatics.
Bioinformatics vs. Computational biology
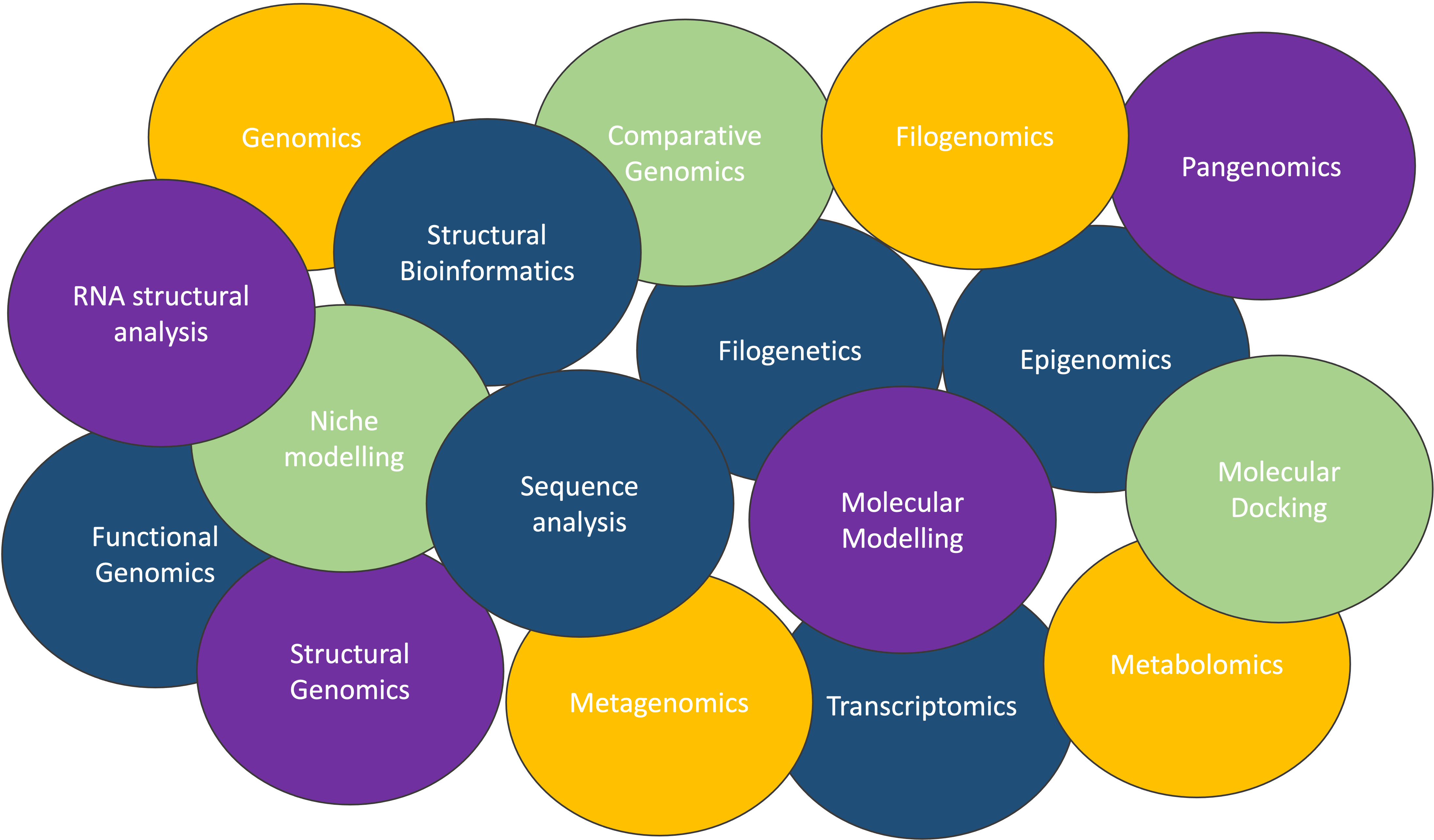
The extent of computational biology
There so many fields on bioinformarics Fig. 1, that sometimes its hard to focus on the fundamentals. But this is also an opportunity to discuss the main aspects and differences across the fields.